The Structure of DNA
Ronald Vale
 Ronald (Ron) Vale
Ronald (Ron) Vale
Ron Vale is a Vice President at Howard Hughes Medical Institute & Executive Director of the Janelia Research Campus where he studies molecular motors, the cytoskeleton and signaling. Ron received his PhD in Neuroscience from Stanford University in 1985. His honors include the Lasker Award in Basic Medical Research, the Shaw Prize in Life Sciences, and the Canada Gairdner Award; he was elected to the US National Academy of Sciences and the National Academy of Medicine. Ron believes in the free dissemination of scientific knowledge throughout the world. He is the founder of iBiology, a nonprofit organization that produces free videos of talks by leading scientists, and the founder and executive producer of The Explorer’s Guide to Biology.
What’s the Big Deal?
The DNA double helix, one of the most influential discoveries in the history of science, revealed how information is stored and replicated in living organisms. Francis Crick and Jim Watson did not exaggerate when they proclaimed that they had found "the secret of life" at the Eagle Pub in 1953. The double helix has since become the foundation of modern biology and its most iconic image. However, DNA is much more than a molecule to be memorized; it is a portal for understanding science and scientific strategy. In this Narrative, we will unravel the DNA helix in our minds and go back to a time when the structure was unknown and DNA was not even known to be the molecule of heredity. What did it take to catapult DNA into the modern era? What seems obvious now was not then. The quest to understand DNA was marked by mistakes by brilliant people, insights by underdogs, clues that lay forsaken, and flashes of brilliance. The path to the double helix also reveals the complementary roles of modeling (Watson and Crick) and experimentation (Rosalind Franklin, Maurice Wilkins, and Florence Bell, an unsung heroine whose early discoveries on DNA are featured here). Springing forward to modern times, we will explore how contemporary bio-engineers use DNA as a building material to create nanoscale devices. The DNA double helix continues to be a source of enormous creativity and inspiration.
Learning Overview
Big Concepts
Complementarity between molecules governs many interactions and reactions in biology; the chemical complementarity between bases (guanine interacting with cytosine, and adenine with thymine) holds two DNA strands together in a double helix and provides simple rules for its replication.
Bio-Dictionary Terms Used
adenosine triphosphate, alpha-helix, amino acid, angstrom, antibody, atomic force microscopy, base, bacterial strain, bacteriophage, Central Dogma, chromosome, CRISPR, gene editing, diploid, DNA cloning, electron microscopy, gamete, genome, gene, germ cell, haploid, hemoglobin, insulin, keratin, meiosis, messenger RNA, mutation, polynucleotide, polypeptide, polysaccharide, protein, reverse transcription, ribosome, RNA, sickle cell anemia, T cell, transcription factor, van der Waals radius, virulence, virus, white blood cell, X-ray, X-ray crystallography
Terms and Concepts Explained
codon, DNA aptamer, DNA chemical structure, DNA origami, DNA polymerase, DNA primase, helicase, hydrogen bond, major and minor groove, nucleoside, nucleotide, Okazaki fragment, phosphodiester bond, purine, pyrimidine, semi-conservative replication, start and stop codon, topoisomerase
Introduction
The model for the DNA double helix in 1953 by Jim Watson and Francis Crick marked the start of a new era of molecular genetics and personalized medicine.
The model provided a surprisingly simple and clear answer to the fundamental question of how the information for life is stored and replicates.
The elucidation of the DNA double helix is a marvelous story of scientific detective work. It also illustrates how the two general scientific approaches of (1) experimental data gathering and (2) model building complement one another to advance our understanding of living organisms.
Part I: Journey to Discovery- The twisting path to the DNA double helix
Gregor Mendel’s experiments with peas in the mid-19th century revealed that different traits are conferred by discrete heritable entities that later were called genes. By the early 20th century, evidence suggested that genes reside on chromosomes, which are fibrous nuclear elements composed of DNA and protein.
DNA (deoxyribose nucleic acid), discovered in 1869 by Friedrich Miescher, is composed of four bases (guanine, cytosine, adenine, thymine). The bases are connected to a sugar (deoxyribose), and sugars are interconnected through phosphate linkages to form a long, unbranched chain.
For much of the first half of the 20th century, the chemical agent responsible for heredity was not known. Most scientists, however, believed that genes were composed of proteins, not DNA. DNA seemed too simple (only 4 different bases) compared to proteins (20 different amino acids) to encode the complex information of life.
Two important experiments, one by Avery, MacLeod, and McCarty (1944) and the other by Hershey and Chase (1952), provided compelling evidence that DNA, and not protein, is the chemical agent responsible for heredity.
The first important breakthrough in understanding the structure of DNA was made in 1938 by Florence Bell, a graduate student working with William Astbury in Leeds, England. Bell found a method to align fibers of DNA and then took X-ray photographs of the fibers. Her results (14 years before the famous photograph of Rosalind Franklin) revealed that DNA has a regular structure and that the bases are stacked at regular 3.4 angstrom intervals.
Another important clue was found by Edwin Chargaff who determined the ratios of the four bases in DNA. He found that guanine and cytosine are present at a 1:1 ratio, as are adenine and thymine. The implication of this finding was not understood at first.
Several scientists were chasing after the DNA structure (structure meaning understanding how the DNA chain adopts a three-dimensional shape). Watson and Crick, two young scientists at the Cavendish Laboratory in Cambridge, England, wanted to make a model, effectively a "guess" or hypothesis of what DNA might look like based upon a minimal amount of data. Linus Pauling, a famous senior scientist at Caltech, was doing the same. Rosalind Franklin and Maurice Wilkins, like Florence Bell previously, were taking X-ray photographs and trying to determine the structure using more information from their experimental work.
To make a model of DNA, you need to ascertain (1) how many chains of DNA are present in the biological form of DNA, and, if more than one chain, (2) whether those chains run in the same or opposite directions, (3) whether they pack straight or twist around one another, and (4) how the chains interact.
Failed models of DNA preceded the correct one. The first (unpublished) Watson and Crick model and the published Pauling model both proposed an incorrect DNA triple helix.
Rosalind Franklin made a key breakthrough by preparing a pure and "biologically relevant" form of DNA (B-form DNA) and generated her famous X-ray photograph: photograph 51.
When Jim Watson saw photograph 51 (a half-year after it was produced), he immediately knew that DNA was a helix, thanks to earlier theoretical work by Francis Crick. This insight triggered an intense episode of model building by Watson and Crick.
Crick also found a clue in the X-ray data that revealed that DNA was composed of two strands oriented in opposite directions. It was a double helix.
A key eureka moment was Watson’s realization that guanine pairs with cytosine and adenine pairs with thymine at the center of the double helix. This insight explained how the two DNA strands are held together, how the double helix can maintain a constant width regardless of its nucleotide sequence, and most importantly, how DNA can replicate.
The close collaboration between Watson and Crick and their combined insights allowed their model for the DNA double helix to come together in just 1 month.
The DNA double helix was a model that was supported with some but not extensive data and many scientists remained skeptical. The experiment by Meselson and Stahl in 1958 showed that DNA replicates just as Watson and Crick proposed and led to the general acceptance of the Watson–Crick model.
Watson, Crick, and Wilkins won the Nobel Prize in 1962. Franklin died of cancer in 1958.
Part II: Knowledge Overview - The Structure And Replication Of DNA
DNA (deoxyribonucleic acid) stores information that can be read out to make RNAs and ultimately proteins. DNA can be copied, allowing life to propagate. DNA can mutate, allowing biological variation.
The building blocks of DNA are nucleotides, small organic compounds consisting of one of four bases attached to a sugar (deoxyribose) and a phosphate.
DNA in a chromosome is a single long chain of millions of connected nucleotides. The nucleotides are connected through phosphodiester bonds, in which one phosphate is linked to the 5′ carbon of one deoxyribose and to the 3′ carbon of another deoxyribose. The asymmetry of 5′ and 3′ phosphodiester bonds confers a polarity to the DNA chain.
The most common biological form of DNA is the "B-form" (see Journey to Discovery ), which is a double helix in which the two chains align in opposite directions. It is a right-handed helix that repeats its twist every ∼10 nucleotides. The phosphate-sugar groups constitute the backbone of the helix; the bases, which lie perpendicular to the long axis, are spaced at 3.4 angstrom intervals.
The DNA double helix has two grooves: the major groove and minor groove. Proteins can bind to these grooves, most commonly, the major groove where proteins can recognize a particular sequence of bases.
The DNA strands are held together by two types of interaction forces: hydrogen bonds between bases (G with C and A with T), which provide specificity, and vertical pi-stacking interactions between the planar bases along each strand.
DNA replication, as first realized by Watson and Crick, makes use of the complementary base pairing (G with C and A with T). Each strand contains the necessary information to specify the construction of its partner strand.
DNA replication in cells involves first untwisting and separating the strands by enzymes (topoisomerase and helicase, respectively) that are powered by energy from ATP hydrolysis. The junction where the double helix bifurcates into two single strands is called the "replication fork." Single-strand binding proteins prevent the separated strands from reannealing (snapping back together).
An enzyme called DNA polymerase incorporates new nucleotides onto the 3′ end of a "primed template." The primed template is made by an enzyme called "DNA primase," which makes a short complimentary copy of a single DNA strand using RNA (creating a temporary RNA-DNA duplex).
One of the two separated DNA strands, the "leading strand" is relatively easy to copy; the 3′ end of the primed template is pointing toward the replication fork. DNA polymerase resides at the replication fork and synthesizes a complementary copy of the leading strand.
The copying of the "lagging strand" is more complicated. Its 3′ end is pointing away from the direction in which the replication fork is unwinding the DNA double helix. To copy the lagging strand, (1) DNA primase creates short RNA-DNA duplexes, (2) DNA polymerase uses the 3′ end of the RNA/DNA duplex to synthesize complementary DNA in the direction away from the replication fork, (3) the RNA in the primed templates is degraded and DNA polymerase fills in the gaps, and (4) discontinuities in the copied DNA are connected by DNA ligase, generating a continuous chain of phosphodiester bonds.
The sequence of bases in DNA contains a code that can be read out to build proteins. DNA employs a "triplet" code (groups of three bases, called codons). One of the 64 possible codons specifies the "start," the first amino acid of a protein (the amino acid methionine). Three codons do not encode amino acids and serve as "stop" signals that terminate the addition of amino acids to the protein. The remaining 60 of the 64 possible triplets are used to specify the other 19 amino acids; many amino acids are redundantly specified by more than one codon.
Part III: Frontiers - DNA Nanotechnology
The natural topologies of chromosomes are simple: either lines or circles. However, scientists realized that the rules of Watson–Crick base pairing could be used to design more complicated shapes of DNA. DNA nanotechnology is a field that builds small devices from DNA.
DNA origami is a technique that creates 2D or 3D DNA structures that are ~1/10,000,000th of a meter in size. It uses a long "scaffold" of single-stranded DNA and hundreds of short DNA sequences (oligonucleotides) called "staples."
Each staple hybridizes (forms Watson–Crick base pairs) with two or more regions of the scaffold, forming double helices that are connected through "branch points." The originally floppy single-stranded scaffold now folds into well-packed helices that form a distinct shape.
The steps involved in making DNA origami involve (1) computer-based design of the staples, (2) chemical synthesis of these oligonucleotides, (3) hybridization of the components (formation of Watson–Crick base pairs), and (4) examination of the final structure by electron or atomic force microscopy.
Shawn Douglas and co-workers used DNA origami to create a "nanorobot" to attack cancer cells.
The DNA nanorobot has two curved halves that can be closed to create a barrel-shaped cylinder. Inside the barrel are antibodies that inhibit the growth of cells.
The nanorobot is designed to keep the barrel closed and the antibodies hidden until it detects certain "signature" molecules on a cancer cell using DNA sequences called aptamers.
The investigators found that the DNA nanorobot inhibited the proliferation of cancer cells in tissue culture.
Closing Thoughts
The DNA double helix discovery marked the beginning of an explosive era of molecular biology. While much has been learned about the functions of DNA, many mysteries still remain.
Watson and Crick succeeded in deducing the structure of DNA because of their passion to solve the problem and their ability to collaborate well together. They were as complementary as the two strands of a DNA double helix.
Watson’s and Crick’s success relied upon experimental X-ray data on DNA, first produced by Florence Bell and then extended by Rosalind Franklin. Bell and Franklin pursued science in an era when it was difficult for women to achieve the same level of independence and recognition in science as men. The story of DNA reminds us today of the importance of ensuring that the scientific profession is fair and welcoming to all races and genders.
Guided Papers
Watson, J.D. and Crick, F.H.C. A structure for deoxyribose nucleic acid. Nature 1953;171:737–738.
This classic paper showed that DNA has a double helix structure, importantly revealing an elegant and simple mechanism for replication and the inheritance of genes. This research led to Watson and Crick receiving their Nobel prizes in 1962.
DownloadWatson, J.D. and Crick, F.H.C. Genetic implications of the structure of deoxyribonucleic acid. Nature 1953;171:964–967.
This classic paper proposes that the precise sequence of DNA nucleotides is the code which carries genetic information, and that each strand of the DNA double helix can serve as a template to create a new copy, thus allowing the genetic information to passed along from one cell or organism to its progeny. With the previous paper, this research led to Watson and Crick receiving their Nobel prizes in 1962.
DownloadDouglas, S.W., Bachelet, I., and Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012;335:831–834.
This paper describes the design and use of DNA nanorobots to transport molecules to cells, by selectively interacting with cells to deliver signaling molecules to cell surfaces.
DownloadIntroduction
How does life propagate? How is information stored? Is there a code for life? Before 1953, answers to these fundamental questions about life remained mysterious.
Then in 1953, the DNA double helix appeared on the scene, a discovery published by two young "nobodies" at the time- Jim Watson and Francis Crick. The structure of DNA that they envisioned was beautiful; two strands embracing and twisting around one another. The DNA double helix has since become the most recognizable and iconic image in biology; its graceful beauty has even inspired artists and architects. However, for biologists, this molecular spiral staircase was not only beautiful, but packed with meaning. The rules for making a DNA double helix answered the basic enigma of how life’s information is stored and copied. The answer was far simpler and more elegant than any scheme that scientists had imagined before.
The DNA double helix has had a powerful influence on biology, arguably equal to the influence of the theory of evolution by Charles Darwin. The DNA double helix initiated the quest to determine how the DNA sequence instructs the production of proteins. Once this code was solved, scientists developed sequencing technology to read the code. Today, the sequence of the six billion letters of your DNA can be determined for $600. This technological advance is ushering in a new era of personalized medicine, which can reveal your ancestry and propensity for developing certain diseases.
The simplicity of the DNA double helix also beckoned engineers to find ways to manipulate it. First, scientists found ways to cut, paste, and amplify DNA, which is called DNA cloning (see Key Experiment by Chalfie). Scientists then found ways of transferring DNA from one organism to another (see the Narrative on Plant Genetics by Ronald). These tools provide much of the foundation of the modern biotechnology industry. Most recently, scientists developed tools to rewrite the sequence of DNA in a genome, which is equivalent to editing a single letter in a massive encyclopedia. You can read about this new CRISPR/Cas technology in the Key Experiment by Doudna and the Narrative by Barrangou. We certainly have not reached the end. Where will the DNA double helix take us next?
Most people have learned about DNA in school and heard about Watson and Crick. But do you know what they discovered?
Explorer’s Question: What did Jim Watson and Francis Crick do?
A. Discovered DNA and solved its structure.
B. Showed that DNA is the hereditary material.
C. Made a model for the structure of DNA without performing any experiment.
D. Performed a key experiment that shows how DNA replicates.
E. Solved the genetic code for DNA.
F. All of the above.
Answer: C
Even if you knew the answer to the above quiz, we will take a much deeper dive into the story of DNA in the Journey to Discovery, examining the detective work that underlies a scientific discovery. How in the world did someone figure out the three-dimensional shape of DNA? And why was the answer so interesting? My hope is that you will be able to explain the answers to these questions to a fellow student, friend, or relative, and take them on a scientific journey.
The story of the DNA double helix reveals the humanity behind science. The story had a triumphant ending for Watson and Crick. But the history of DNA is also littered with mistakes, stubbornness, and missed opportunities. It may surprise you to learn that scientists thought that DNA was a "boring" molecule for many decades. Alas, scientists are human. Few scientific studies hit the bull’s eye right away, and mistakes are part of the process of tackling the unknown.
The Journey to Discovery also highlights the interplay between model building and experimental data gathering, two important and complementary strategies that continue to drive biological discovery today. Jim Watson and Francis Crick were the "modelers"; they, in fact, never did an experiment themselves on DNA (although they performed experiments for other projects). The experimentalists in our story are Florence Bell, Rosalind Franklin, Maurice Wilkins, and Erwin Chargaff; they provided the data that Watson and Crick needed to build their model. Of these individuals, Florence Bell is virtually unknown, although I hope that you will enjoy discovering, as I did, how her work provided a foundation for understanding DNA structure. I also hope to convey the difference between a model, which is a hypothesis, and a fact. In 1953, the Watson and Crick DNA double helix was a hypothesis, and not a fact, and many scientists were not convinced. A good model, however, makes predictions that guide the next round of experiments. The double helix model triggered Meselson and Stahl to do their Key Experiment, which provided critical validation and broader acceptance of the double helix.
In the Knowledge Overview, we will take a look at the DNA double helix more closely and examine how complementary base pairing (guanine with cytosine and adenine with thymine) guides the shape and replication of the helix. Moving to the Frontiers section, we will explore how the rules of DNA base pairing are being exploited by scientists today to design and build three-dimensional, DNA-based robots and other devices that are less than a millionth of an inch in size. This new field of DNA nanotechnology is just one of many examples in which the DNA double helix continues to inspire creativity in research and biotechnology.
In the Closing Thoughts, I will reflect upon personal and professional insights that might be derived from the story of the DNA double helix. One key point, applicable to any endeavor in life, is the importance of collaboration, as illustrated by the incredible partnership between Watson and Crick. The second point is the importance of scientific independence and recognition, which was more accessible to men than women in the mid-20th century. The experiences of Florence Bell and Rosalind Franklin remind us that we still need to be vigilant in making the scientific profession gender inclusive. Whether you are an aspiring scientist, entrepreneur, sociologist, politician or artist, there are interesting lessons from the twists and turns of the DNA double helix.
Part I: Journey to Discovery —
The Twisting Path to the DNA Double Helix
[Defining the chemical nature of the gene] "is one of the great urgent tasks of humanity. Once we have accomplished it, the limits of our horizons widen beyond imagination, glimpsed at last what we are."
- Florence Bell, from her PhD thesis (1939). See references.
The Problem
How are traits passed from parents to offspring? The first breakthrough on this problem was made by Gregor Mendel, a 19th-century monk who studied peas. Mendel showed that traits are inherited as discrete units, which later became known as genes (see Narrative on Inheritance by Tilghman). The gene became a very important concept in biology, and by the early 20th century, it was known that genes could:
1) Store information. Previous work had demonstrated that one gene contains information that instructs the production of one protein.
2) Replicate. A copying mechanism must exist that allows genetic information to be propagated from one generation to the next.
3) Mutate. Occasional changes to genes produce offspring with distinct traits from their parents. This variation is the driver of evolution through natural selection (see Narrative on Mutations by Doug Koshland).
But what is a gene? What is it made of? What does it look like? If one knew the physical/chemical nature of gene, perhaps one could understand how genes store information, replicate, and mutate.
Evidence at the turn of the 20th century suggested that genes reside on chromosomes (see Dig Deeper 1). Chromosomes are made up of both DNA and proteins. Which was the gene? For much of the first half of the 20th century, scientists thought that genes were made of protein, not DNA, for the following reasons:
1) Polypeptides (another name for proteins) are linear chains composed of 20 different building blocks- the amino acids. DNA, on the other hand, is composed of only 4 different building blocks—the bases. Because proteins are more complex than DNA, polypeptides were viewed as a better bet for storing the complex information of life.
2) DNA was thought to be a monotonous, repetitive string of nucleotides, which would not be useful for information storage. Phoebus Levene promoted this view with his "tetranucleotide hypothesis," which postulated that DNA’s four bases are present in equal amounts and repeat over-and-over again along the chromosome in a fixed pattern. This hypothesis turned out to be completely wrong, but it was influential in its time.
As a result of this frame of thinking, many scientists thought DNA was boring for approximately the first 80 years following its discovery. DNA stands as an example of why eliminating incorrect ideas is often as important as creating new ones.
In our Journey to Discovery, we will encounter two famous experiments, one by Avery–MacLeod–McCarty and another by Hershey–Chase, which together overturned the idea that proteins are the molecule of heredity and pointed the finger squarely at DNA (explained in Clue 3 below). The tetranucleotide hypothesis was finally overturned by the data described below in Clue 4.
Yet, even with these wrongs corrected by 1952, no one could imagine how DNA functions in heredity. Even the notion that DNA contains a code, a basic fact about DNA that most people know today, was unappreciated at the time. Max Delbruck (a well-known scientist featured in Koshland’s Narrative on Mutations and the Meselson-Stahl Key Experiment) said:
"Nobody, absolutely nobody, until the day of the Watson-Crick structure, had thought that the specificity might be carried in this exceedingly simple way, by a sequence, by a code."
- Quote from The Eighth Day of Creation (see reference list)
Our Journey to Discovery ends with the model of the DNA double helix proposed by Jim Watson and Francis Crick. The structure unexpectedly suggested an elegant mechanism for the replication and inheritance of genes, solving the puzzle posed by Mendel a century earlier. The once-boring DNA molecule has been in the limelight ever since.
Clues
Clue 1: The Chemistry of DNA
In 1869, Johann Friedrich Miescher was studying the chemical nature of "pus," which he obtained from discarded surgical bandages from a local surgery clinic. His studies of pus led to the discovery of a new substance, which he named "nuclein" that was acidic, rich in phosphorus, and lacked sulfur, differentiating it from proteins. Soon after, he found an even better source of nuclein—salmon sperm. What Miescher discovered is now known as deoxyribonucleic acid or DNA.
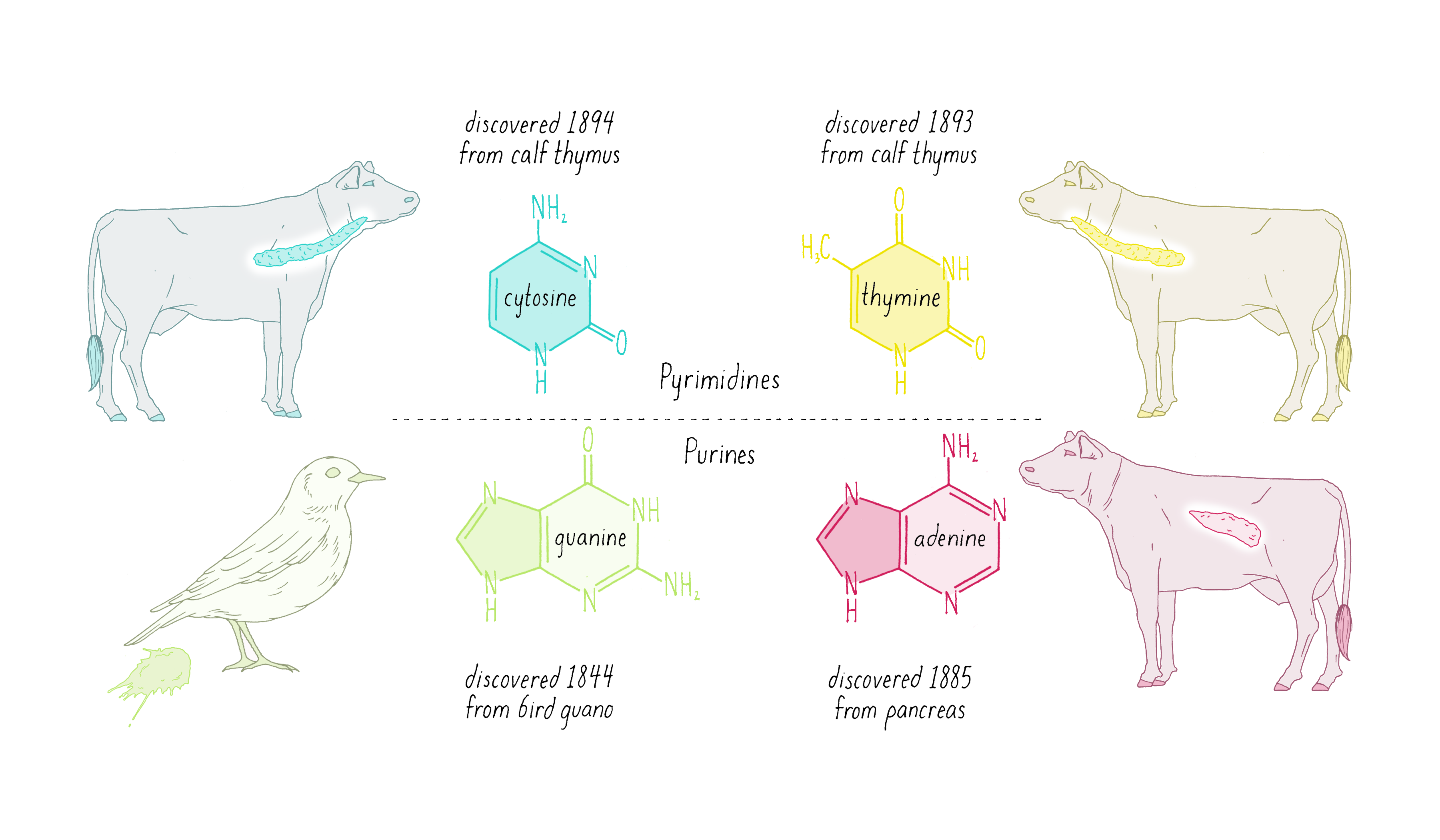
Nuclein was discovered to be composed of four distinct chemical units called bases—adenine, guanine, thymine, cytosine (Figure 1). Guanine (abbreviated as "G") was first found in 1844 in the excrement of birds (called "guano"). Adenine (abbreviated as "A") was isolated in 1885 from beef pancreas. Guanine and adenine are similar to one another (a hexagon of carbons joined to a pentagon with a common side) and are called "purines." The other DNA bases, thymine (T) and cytosine (C) were first isolated from calf thymus glands. Thymine and cytosine (hexagons) are called pyrimidines.
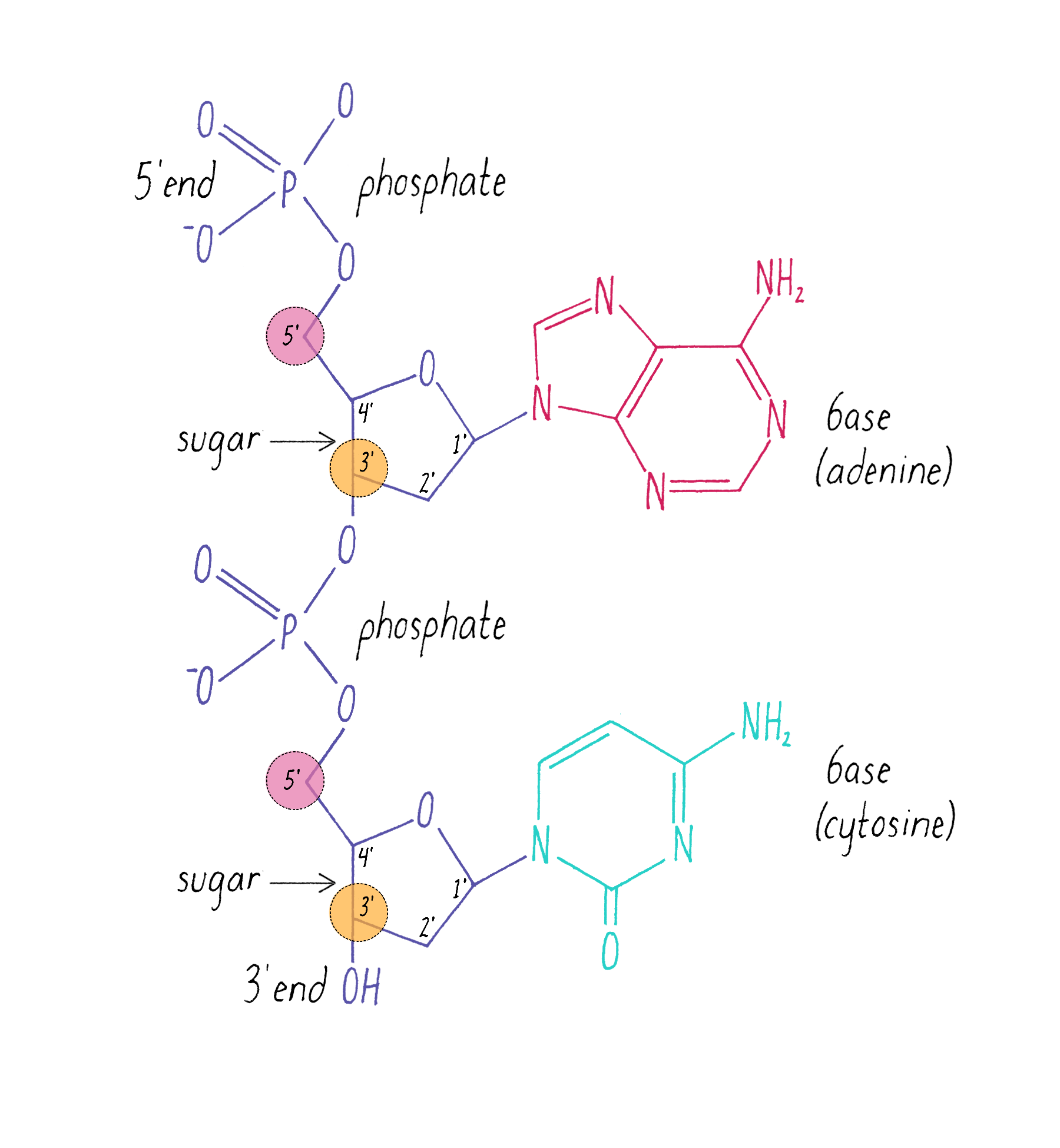
The chemical structure of DNA was further elucidated through the work of several scientists, most notably Alexander Todd who won a Nobel Prize for his work. More details will be presented in the Knowledge Overview, but the fundamentals are presented here since they pertain to the discovery of the DNA structure. The basic building blocks of DNA are not the bases alone but are nucleotides. A nucleotide is a base (one of the four) connected to a sugar (deoxyribose for DNA), which in turn is connected to a phosphate (Figure 2). DNA is a long polymer of nucleotides and is sometimes called a polynucleotide (many nucleotides). The nucleotides are connected in a head-to-tail fashion by bonds formed between one phosphate and two adjacent sugars; one of the bonds connects the phosphate to what is referred to as the 5′ carbon on one sugar, and the other bond is formed with the 3′ carbon on another sugar. Thus, the phosphates and sugars create a continuous backbone of the DNA molecule with the bases sticking out at regular intervals. The DNA chain has directionality, as defined by the way in which the 5′ and 3′ carbons on the sugars are pointing (Figure 2).
Clue 2: The First X-ray Structure of DNA by Florence Bell
Rosalind Franklin produced a famous X-ray photograph of DNA in 1952, which we will discuss soon. However, the first X-ray photograph of DNA was obtained 14 years earlier at the University of Leeds by a young graduate student—Florence Bell.
Born in London in 1913, Florence Bell (Figure 3) studied chemistry and physics in college and was taught X-ray crystallography by two leading scientific experts. Well-trained, she then moved to Leeds in 1937 to begin Ph.D. studies with William Astbury, another well-known X-ray crystallographer. Bell received her Ph.D. in 1939; her thesis was titled "X-ray and related studies of the structure of the proteins and nucleic acids" (see references). Bell produced an extraordinary PhD, which has been largely overlooked. It shouldn’t be.

Bell’s research involved shining a beam of X-rays through aligned biological fibers, such as DNA. The fibers scatter some of the X-rays, which are then captured as an image using X-ray sensitive film. A fuzzy, diffused image on the film indicates a lack of a regular order of molecules in the fiber. However, a pattern of spots or lines reflects an underlying order of atoms in the fiber. As will be discussed later, information on the arrangement of atoms in the fiber potentially can be deduced from the X-ray pattern. However, Bell was working in the very early days of using X-rays to probe the structures of molecules. X-rays had been used to determine the arrangements of atoms in simple salt crystals, but the structures of biological molecules, which are more complicated than simple salts, had not yet been solved by this method.
Bell passed X-ray beams through an extraordinary number of biological substances, which sound like a witch’s brew—tendons of frog toes, jellyfish, human hair, the ribs of shark fins, etc. These samples are made of proteins. However, Bell also looked at DNA from various sources. One sample of DNA (from calf thymus) produced interesting results.
If you prepared DNA from strawberries in primary or middle school, you might remember DNA as a gooey blob. As discussed above, a blob of disorganized molecules will produce a fuzzy X-ray image devoid of information. Thus, Bell first had to find a way to align the DNA molecules. She did so by drying DNA onto a frame that could be slowly stretched. The stretching helped to align the DNA molecules in the same direction, thus creating some degree of order.
Bell passed the X-ray beam through her calf thymus DNA fibers and the scattered X-rays produced an image on the photographic film (Figure 4). It was a bit smeary, but there was a pattern. Just observing a pattern was a big result; it was the first indication that DNA has some type of molecular order. This was not a foregone conclusion. For example, X-rays would not produce any pattern if passed through a preparation of RNA (another polynucleotide) or polysaccharides (polymers of sugars); these polymers can adopt numerous 3D structures and thus produce irregular fibers.
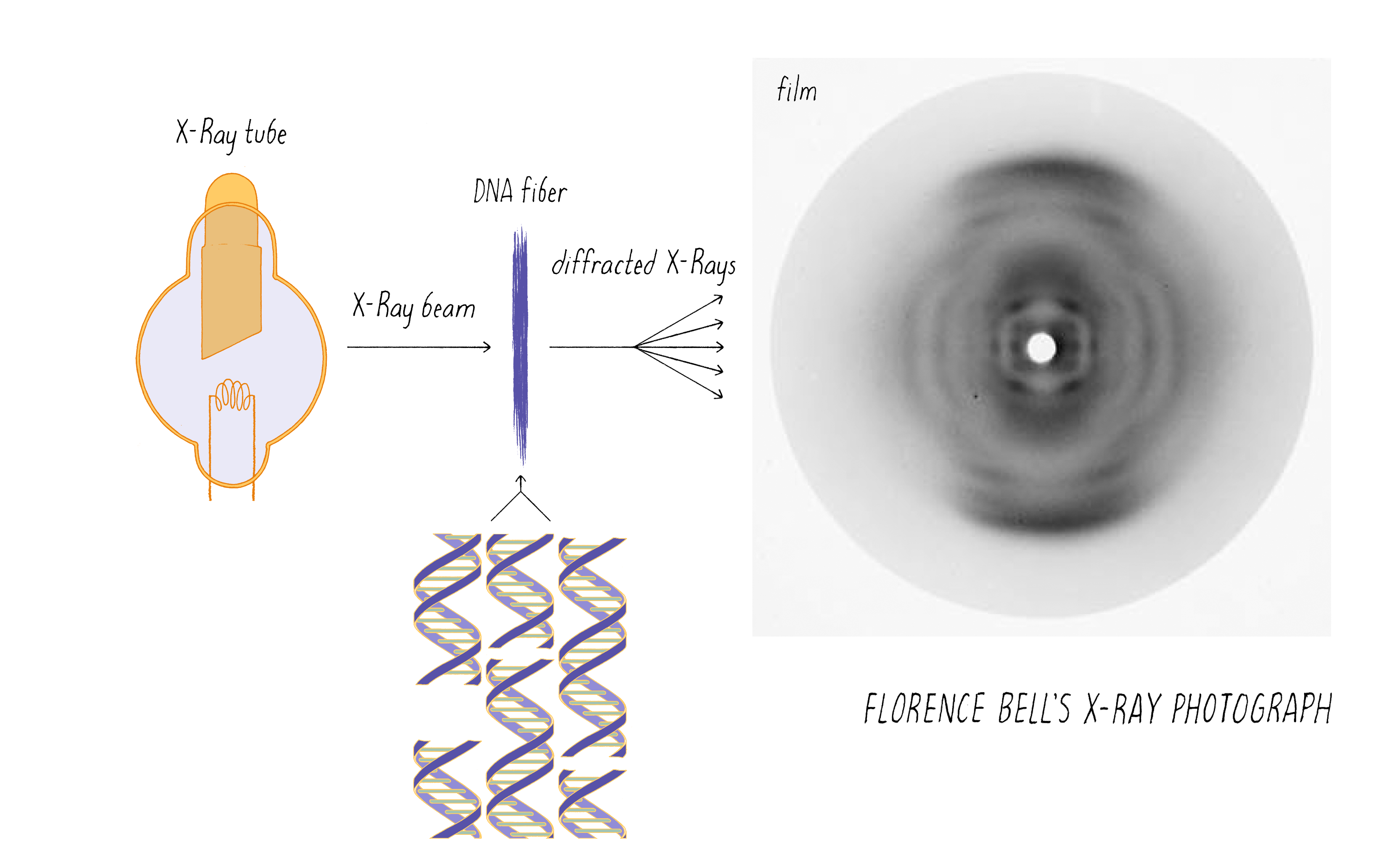
Bell learned several things about DNA from her X-ray photographs. First, the big semi-circular band at the very top and bottom (which is called a "reflection" in the lingo of X-ray diffraction) revealed that some element in DNA repeats at regular intervals of 3.4 angstroms (angstrom, abbreviated with the symbol "Å," is 10−10 meters; one-tenth of a nanometer). She verified this periodicity in DNA obtained from viruses and pancreas, thus demonstrating that it must be a universal property of DNA. From other information in the photograph, she determined the width of a DNA molecule as 18.1 Å (close to the currently accepted value of 20 Å). From these data, Bell produced a model in her thesis (Figure 5) showing that:
1) DNA is a long column that forms a rigid structure.
2) The bases lie perpendicular to the axis of the column and stack every ~3.4 Å. (This was a key parameter in the Watson and Crick model, as we will soon see).
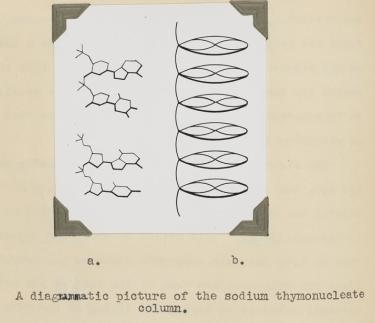
That DNA is a rigid column with a width of ∼20 Å and its bases stack every 3.4 Å is correct. However, Bell did not reach the conclusion that DNA is a double helix. Her preparation of DNA (like the early ones of Franklin) was a mixture of two forms of DNA, which complicated matters and blurred the image. Even if she had the "right" photograph, the theory to interpret an X-ray pattern of helical structure did not exist at the time.
While the model for DNA shown in Figure 5 is not fully correct, Bell’s pioneering work in 1938 represented a significant achievement, a full 15 years before the famous DNA papers that appeared in 1953. Bell was the first person to show that DNA has an orderly underlying structure. Her work indicated that obtaining a structure for DNA was a doable undertaking, and attracted others (Franklin, Wilkins) to take the problem to the next level. In fact, Franklin continued to study thymus DNA, the same source of DNA that Bell found worked best in her structural studies. Science proceeds in steps, which lay the necessary foundation for big advances.
Many years later in 1951, Bell’s advisor William Astbury produced a better X-ray photograph of DNA, indeed one similar to the famous photograph produced by Rosalind Franklin (discussed below). However, Astbury never published or presented this result publicly; it was likely that he did not grasp its significance. If it was published and accessible to Linus Pauling and Francis Crick, the history of the DNA double helix would very likely have been altered.
Could Florence Bell have cracked the problem of the DNA double helix? We cannot know the answer. World War II disrupted her scientific work (as was true for Francis Crick’s, as we will see later). Astbury tried to intervene in her recruitment by the British War Office, stating that he "could hardly carry on without her help." But Bell joined the war effort as a radio operator in the Women’s Auxiliary Air Force. She then married an American serviceman, came to the USA and left science for good. We will return to Florence Bell in the Closing Thoughts.
Clue 3: The Avery-MacLeod-McCarty and the Hershey-Chase experiments point to DNA, and not protein, as the material of heredity
As discussed earlier, chromosomes are composed of proteins and DNA. But which stores the information of the gene? Proteins were favored but an experiment was needed to settle this issue. That experiment was done in 1944 by Oswald Avery, Collin MacLeod, and Maclyn McCarty.
Avery, MacLeod, and McCarty followed up on a remarkable observation made by Frederick Griffith in 1928. Griffith was studying two related strains of the bacteria Streptococcus pneumonia—one strain that was virulent and caused pneumonia when injected into mice and another stain that was non-virulent (did not cause disease). The difference between the two strains was that the virulent strain had a coating that protected it from destruction by the immune system. Griffith did an experiment in which he killed virulent bacteria with heat and co-injected these dead bacteria with live non-virulent bacteria in mice. What possessed him to do this experiment is not clear from my reading, but the results were astonishing. Griffith found that some of the non-virulent bacteria were permanently "transformed" into virulent bacteria that would produce pneumonia (Figure 6). This result suggested that genes (in this case, ones conferring the protective coat that would make the bacteria virulent) were transmissible between bacteria, even, incredibly, from dead bacteria to living ones. This was the first experimental demonstration of gene transfer, a method that powers today’s biotechnology industry. Diabetic patients, for example, are treated with human insulin made by bacteria transformed with the human insulin gene.
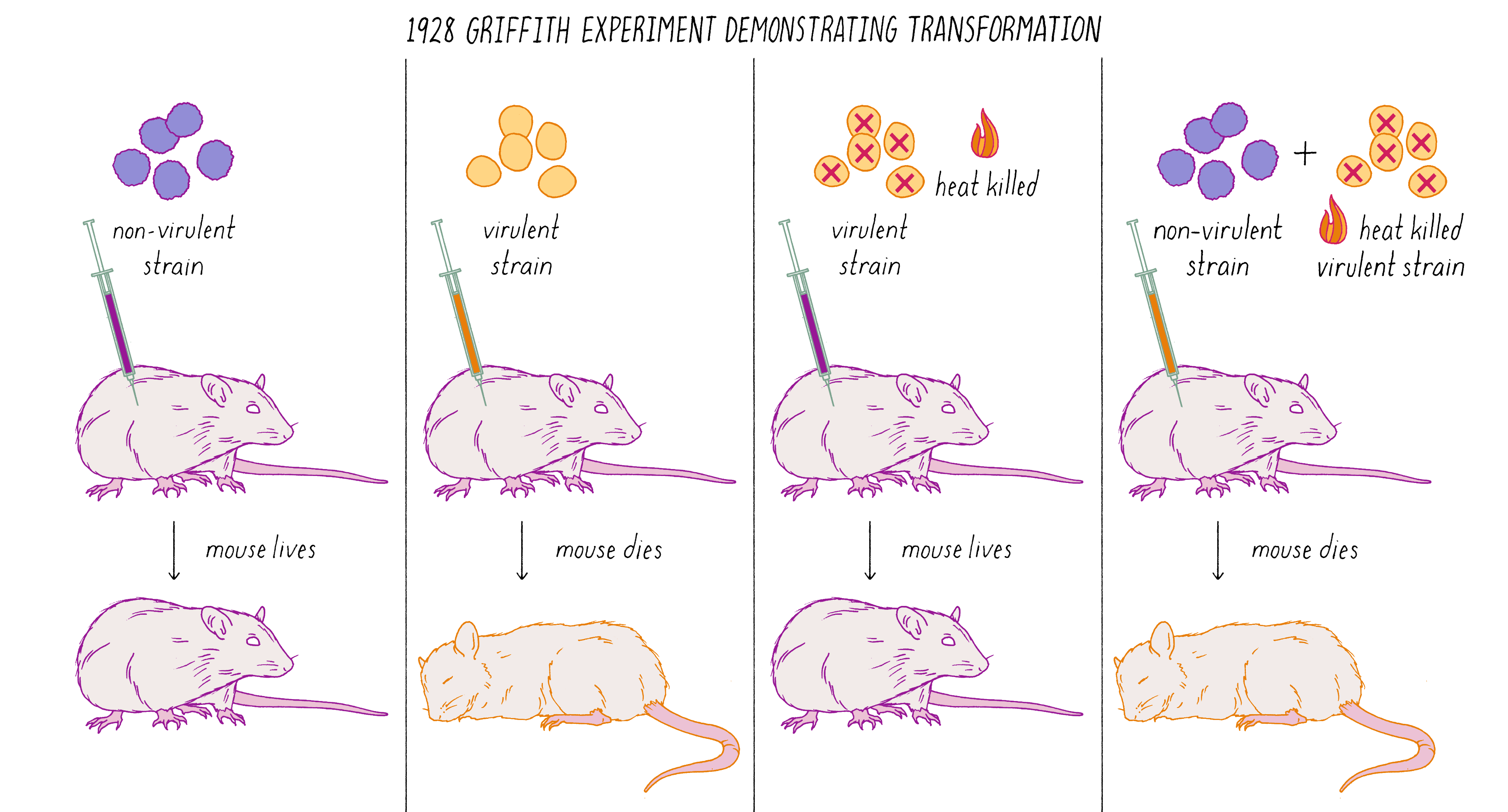
What was the chemical agent that was transferred between the bacteria during genetic "transformation?" Avery, MacLeod, and McCarty found that DNA, and not protein, was the responsible agent. They argued that genes were made of DNA. This result was surprising and stood against the dogma of the time. You can learn more about the Avery, MacLeod, and McCarty experiment in Video 1.
Many scientists, however, resisted the conclusion that genes are made of DNA. The dissenting scientists thought there might have been some remaining proteins that contaminated the DNA preparation. Despite the elegance of the Avery, MacLeod, and McCarty experiment, the chemical nature of the gene remained unsettled in many people’s minds.
Long-standing ideas are often hard to dethrone. However, wrong ideas will eventually crumble under the weight of scientific evidence. The next evidence in favor of DNA came 8 years later (1952) in an experiment performed by Alfred Hershey and Martha Chase. Hershey and Chase studied the replication of viruses, called bacteriophage (phage for short), that infect bacteria. Phage land on the surface of the bacteria and inject a molecule inside. Then many new phage are produced inside the bacteria. The molecule that was injected into bacteria surely must be responsible for heredity since it possesses information to specify the production of new phage. Hershey and Chase discovered that the DNA, and not protein, was injected into the bacteria by the phage. To learn how Hershey and Chase did their experiment, see Video 2.
Since both the Avery–MacLeod–McCarty and the Hershey–Chase experiments, using very different methods, both came to the same conclusion, the scientific community began more widely to embrace the notion that genes are made of DNA.
In our next clue, we will encounter a scientist who immediately recognized the importance of the Avery–MacLeod–McCarty experiment and shifted his research program from proteins to DNA. That individual was Erwin Chargaff.
Clue 4: Chargaff’s Findings
Erwin Chargaff was a biochemist who tackled a simple question: how much of each of the four bases is present in DNA? The tetranucleotide hypothesis, proposed by Phoebus Levene, stated that bases should be present in equal proportions (one-quarter each). Chargaff chemically analyzed DNA to see if this was true or not. Chargaff’s results are summarized in Figure 7.
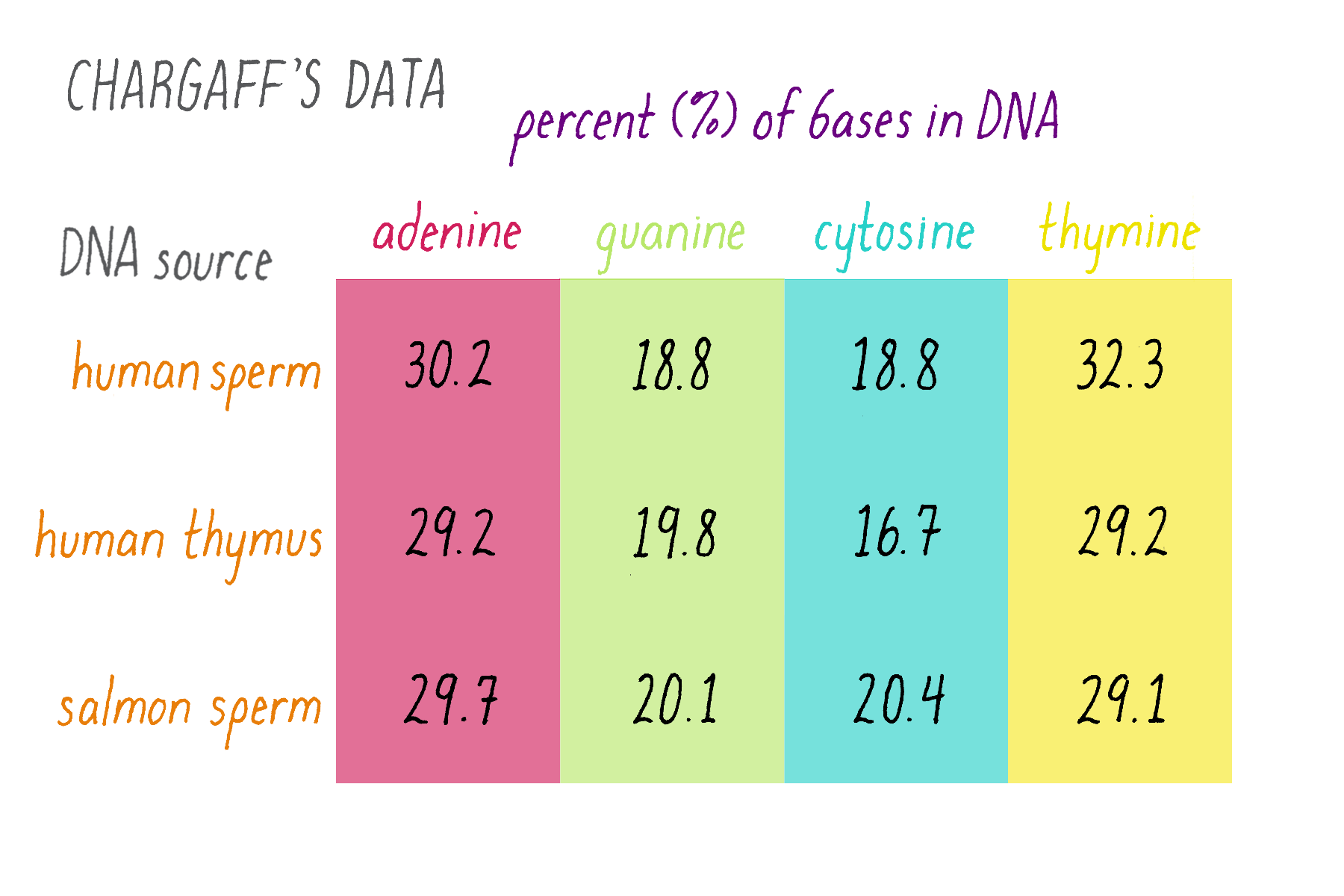
Explorer’s Question: What do these results say about the "tetranucleotide hypothesis?"
Answer: The tetranucleotide hypothesis proposed that the bases repeat in a regular manner along the DNA, which predicts equal (25%) amounts of the four bases. Chargaff’s data, however, showed that this is not the case. In other data, Chargaff also found that the abundance of the bases differs to some extent in different organisms (e.g., yeast, bacteria, human), whereas the tetranucleotide hypothesis envisioned a similar percentage of the bases in all organisms.
Explorer’s Question: Do you notice any similarities in the relative amounts of the nucleotide bases?
Answer: The percentages of adenine and thymine are similar. So are the percentages of guanine and cytosine (although they differ somewhat in the human thymus, likely due to experimental variation or error).
Erwin Chargaff’s finding of equal percentages of adenine and thymine, and of guanine and cytosine are now known as "Chargaff’s Rule." However, it seemed less obvious as a "rule" at the time. Chargaff himself seemed unsure of the meaning in his 1950 paper:
"It is noteworthy, though possibly no more than accidental, that in all deoxypentose nucleic acids examined thus far that the molar ratios of total purines to total pyrimidines were not far from 1. More should not be read into these figures."
If one believed in Chargaff’s data, the unitary ratios of G to C and A to T strongly suggested some sort of complementarity. A matching lock and key is an example of structural complementarity. Were G-C and A-T complementary locks and keys?
We will come back to the Chargaff data, as it provided confirmation of the Watson–Crick model of DNA. Clues are easier to see in hindsight and often missed at the time. Even Watson and Crick did not seize upon the meaning of Chargaff’s data early on. However, once they fully realized its meaning, the two strands of DNA snapped together in their minds into a double helix, as we will see shortly.
The Discovery: DNA is a double helix
Figure 8 is an illustration of one of the most famous photographs in biology: Jim Watson and Francis Crick standing in front of their 3-D metal model of the DNA double helix in Cambridge, England in March 1953. The scientific detective work that led to this model unfolded in an amazingly short period of time, approximately 4 weeks. We will explore what happened in this miracle month.
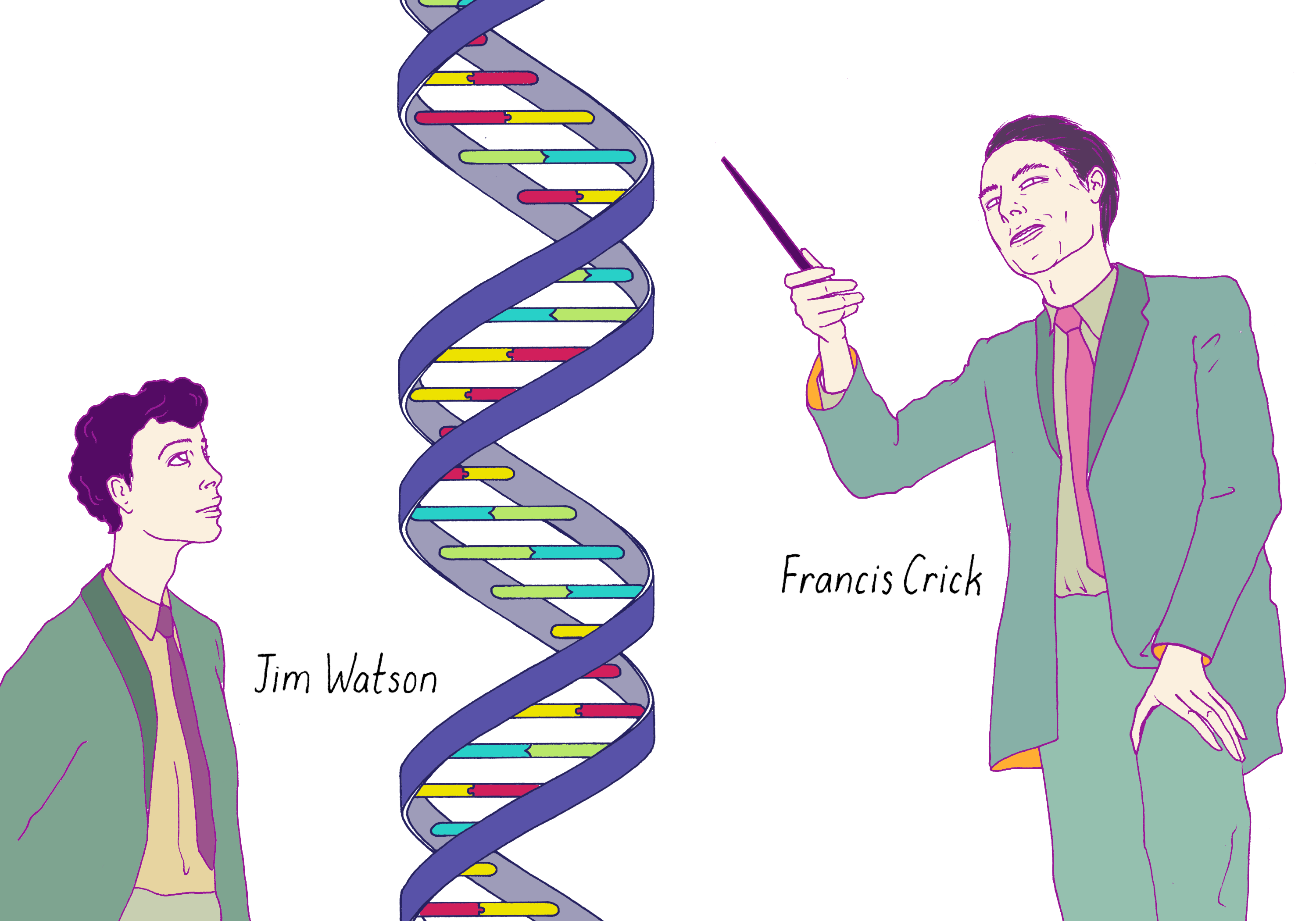
Before we examine how Watson and Crick got to their triumphant moment, let’s introduce the scientists in the story.
The scientists
Linus Pauling: The Favorite
If one was going to place a bet on who would solve the structure of DNA, the smart money would have been placed on Linus Pauling. Pauling, a professor at Caltech, was one of the most famous scientists of the 20th century; he literally "wrote the book" on modern chemistry: The Nature of the Chemical Bond. Pauling also was a pioneer in applying chemistry to biology. In 1949, he correctly postulated that sickle cell anemia is a disease involving a molecular defect in hemoglobin. In 1951, just 2 years prior to the DNA structure, Pauling made another major splash by building an atomic model for the alpha-helix, a common structural motif in proteins. Linus Pauling also articulated the concept of molecular complementarity, a lock-and-key-like mechanism for how two molecules could uniquely interact with one another. In 1948, he stated why complementarity might be relevant to how a gene makes a copy of itself, indeed foreshadowing the big concept behind the DNA double helix:
"If the structure that serves as a template (the gene or virus molecule) consists of, say, two parts, which are themselves complementary in structure, then each of these parts can serve as the mold for the production of a replica of the other part, and the complex of two complementary parts thus can serve as the mold for the production of duplicates of itself."
Linus Pauling, from Molecular Architecture and the Process of Life
Jim Watson and Francis Crick: The Underdogs
Watson and Crick, unknown young scientists working at the Cavendish Laboratory in Cambridge, England, had complementary skills and personalities, which allowed them to come up with the model of the double helix.
Jim Watson: Watson first studied ornithology as an undergraduate at the University of Chicago and later became interested in genetics. Rejected by Caltech and Harvard for graduate school, Watson ended up at U. Indiana training with the superb bacterial geneticist Salvador Luria (Luria is featured in the Koshland Narrative on Mutations). After obtaining his Ph.D. at the age of 22, Watson left for Europe to do postdoctoral studies in Denmark. Upon seeing Maurice Wilkins (discussed below) present his X-ray work on DNA at a scientific meeting, Watson decided that investigating DNA structure with Wilkins would be more exciting than his current work in Copenhagen; however, Wilkins did not invite Watson to join his group. Watson then applied to the Cavendish Laboratory in Cambridge, a leading center for X-ray studies. He arrived in Cambridge in October 1951, 23 years old at the time, where he met Francis Crick. During their first meeting, they both expressed a keen interest in DNA and resolved to work on it together, even though it was not the primary work assigned to them.
Francis Crick: Crick first started Ph.D. studies in physics at University College London, but his work was disrupted when his department had to evacuate to Wales at the start of World War II. Crick then joined the war effort, designing naval mines. After the war was over, Crick returned to science but became interested in how physics could be applied to biology (see also the story of Luria and Delbruck, two trained physicists who turned to biology in the Narrative by Koshland). Crick then entered the Cavendish Laboratory in 1949 for Ph.D. training. When Watson and Crick met in 1951, Crick was 12 years older than Watson but further behind in his training, as he was still a graduate student while Watson was a postdoctoral fellow. While recognized for his tremendous intellect, Crick also could be loud, bold, and sometimes annoying to the senior scientists. To get some respite from Crick, the senior scientists at the Cavendish Laboratory moved Crick and the new recruit Watson to their own room. Would the double helix model have emerged if Watson and Crick had been assigned to work in separate rooms? One cannot say but certainly, their close quarters allowed for constant discussion, which was a key ingredient for their success.
Rosalind Franklin and Maurice Wilkins: The experimentalists who got the data
The information from the X-ray photograph obtained by Florence Bell (described earlier in Clue 2) was insufficient to determine the DNA structure. A next-generation of X-ray crystallographers took up the challenge—Maurice Wilkins and Rosalind Franklin at King’s College, London.
Maurice Wilkins: Wilkins, like Crick, was originally trained as a physicist (obtaining his Ph.D. in 1940) and helped the British and US war efforts in WWII. Wilkins then joined a new Biophysics Unit at King’s College London and began X-ray studies of DNA. He obtained decent X-ray photographs of DNA in 1950, which Watson saw a year later. Wilkins kept close communication with Watson and Crick between 1951–1953. Watson and Crick invited Wilkins to coauthor their DNA structure paper, but Wilkins turned down the offer and published his own X-ray work. Wilkins’ ongoing work on DNA helped to solidify the double helix model; he was a co-recipient of the Nobel Prize with Watson and Crick in 1962.
Rosalind Franklin: Franklin was born in London and attended Cambridge University. Trained originally as a physical chemist, she worked in Paris as a postdoctoral fellow, where she became an accomplished X-ray crystallographer. Franklin received a 3-year fellowship to work on proteins at King’s College London. However, John Randall, the Director of King’s College, reassigned her to work on DNA. Poor communication between Randall, Wilkins, and Franklin led to an acrimonious relationship between Franklin and Wilkins. Wilkins thought that Franklin was assigned to work with him, and Franklin thought that she would be allowed to work independently. We will return to this point in the Closing Thoughts.
The Approaches: Experimentalists and Modelers
The Experimentalists: Maurice Wilkins and Rosalind Franklin
Franklin and Wilkins sought a direct approach to solving the DNA structure using X-ray data. The pattern of spots on the X-ray photograph provides information that, in principle, could be used to reconstruct a 3D image of the DNA molecule.
How does one use X-rays to generate an image of DNA? A camera uses visible light and lenses to recreate images of the macroscopic world. A microscope uses the same principles, with a more powerful lens, to make images of the microscopic world of cells. However, the smaller atomic world cannot be visualized with visible light. Instead, the shorter wavelength of X-rays (∼0.1 nm compared to 400–700 nanometers for visible light) is well matched to the dimensions of atoms. But since it is not possible to make lenses that focus X-rays, direct images of atoms cannot be generated using X-rays. However, unfocused X-rays scattered by the sample can be captured on film, producing a pattern of spots or lines called a diffraction pattern. The diffraction pattern contains information on the 3D arrangement of atoms in a sample. A mathematical treatment (Fourier transform) of the diffraction pattern can recreate an image, thus serving a comparable function to a lens.
Using X-ray diffraction information to reconstruct a detailed 3D model of atoms in a molecule is very common today (if one can get the molecule to form a well-ordered crystal or fiber). However, at the time of Bell, Wilkins, and Franklin, this method was still in its infancy; a structure of a biological molecule as complex as a protein or DNA had not yet been solved at the time. Making matters even more difficult, the mathematical reconstruction of an image had to be calculated by hand in the 1950s, whereas powerful computers perform these tasks today.
The Modelers: Watson and Crick and, separately, Pauling
Watson and Crick wanted to determine the structure of DNA but without the slow and hard work of solving it through a reconstruction of X-ray data. Besides, they were aware that Wilkins and Franklin at King’s College were going down that path and it seemed no point in duplicating their effort. Watson, in particular, was impatient and wanted to solve the puzzle as soon as possible. So Watson and Crick turned to model building. However, other projects occupied most of their time, and they turned to DNA model building sporadically.
Making a model is a process of deduction based upon a limited amount of known information. It is a bit of a game, somewhat like "name that tune". Identifying a song after hearing a minute of music can be relatively easy. But it might be hard to guess from 2 or 3 notes. Florence Bell’s work generated two pieces of information: (1) the distance between bases and (2) the width of the DNA molecule. But that was not enough information to make a correct model. One needed more information, but how much more? Model building also is very dependent on having good data; incorrect data will be misleading and lead to an incorrect model.
Linus Pauling was the quintessential model builder. Pauling’s ability to deduce the structure of the protein alpha-helix through intuition and rules of chemistry inspired Watson and Crick. Watson recounts:
"I soon was taught that Pauling’s accomplishment was a product of common sense, not the result of complicated mathematical reasoning. Equations occasionally crept into his argument, but in most cases, words would have sufficed. The key to Linus’ success was his reliance on the simple laws of structural chemistry. The alpha-helix had not been found by only staring at X-ray pictures; the essential trick, instead, was to ask which atoms like to sit next to each other. In place of pencil and paper, the main working tools were a set of molecular models superficially resembling the toys of preschool children. We could thus see no reason why we should not solve DNA in the same way. All we had to do was to construct a set of molecular models and begin to play- with luck, the structure would be a helix. Any other type of configuration would be much more complicated."
In The Eighth Day of Creation (see reference list)
A model is not a fact; it is a hypothesis for how something works that needs scrutinizing and testing. Sometimes scientists fall in love with beautiful models that are incorrect. How do scientists come to the conclusion that a model is right? This is often a difficult question since several models might be consistent with available experimental data, especially if the data are limited. However, a good model makes new predictions that guide new experiments that will further test whether a model is right or distinguishes between competing models.
At First, Failure
Before we examine how Watson and Crick got to their triumphant moment, let’s examine the menu of choices involved in converting the 2D chemical structure of DNA into a three-dimensional structure:
1) How many chains of DNA are present in a biological DNA molecule? Is it one, two, three, or more?
2) If more than one, how are the chains oriented? Do they run in the same or opposite directions?
3) If more than one chain, how do they interact with one another? Do the chains interact in a planar fashion or do they twist around one another? If twisted, what is the direction of the twist, and over what distance does the twist make a complete turn?
4) If more than one chain, are the phosphates on the inside and the bases on the outside? Or vice versa?
The limited number of parameters in constructing a 3D model might make this seem like an easy puzzle to solve. However, that DNA is a double helix only looks obvious in retrospect. Watson/Crick and Pauling favored the wrong choices first before the correct solution was identified. The DNA story reveals that coming up with the correct solution to an unknown problem is hard. Scientists typically do not gravitate immediately to the right answer; instead, they get there through an iterative process.
Watson and Crick produced their first model in November 1951, just after Watson heard a presentation on the X-ray work on DNA from the scientists at King’s College. The first Watson–Crick model consisted of three DNA strands wrapped in a triple helix with the phosphates at the center and the bases pointing outward. This idea was pleasing at the time. If the bases, the most interesting and variable part of DNA, were placed on the outside, then they could more easily interact with proteins.
Watson and Crick presented their three-helix model to Wilkins and Franklin. It was premature. Watson did not fully digest the X-ray information presented by Franklin, which led to errors in their model. Franklin immediately called attention to those errors, including having the phosphates in the center. It was a humiliating moment, not only for Watson and Crick but for the Cavendish Laboratory overall. The Director of the Cavendish Laboratory, Sir Lawrence Bragg, banned Watson and Crick from working on DNA and indicated that they should leave the structure of DNA for King’s College to solve. There was a long hiatus (over a year) before Watson and Crick returned to working seriously on DNA again.
Meanwhile, Linus Pauling was having his turn at DNA model building at Caltech. Watson and Crick were friends with Linus’ son, Peter Pauling, who was studying at Cambridge at the time. In late January 1953, Linus Pauling eventually produced a manuscript describing his model for DNA, which Watson and Crick saw via Peter.
Pauling’s model (which appeared in print in February 1953) had the same mistake as the first model of Watson and Crick. It too was a three-stranded helix with the phosphates at the center (Figure 9).
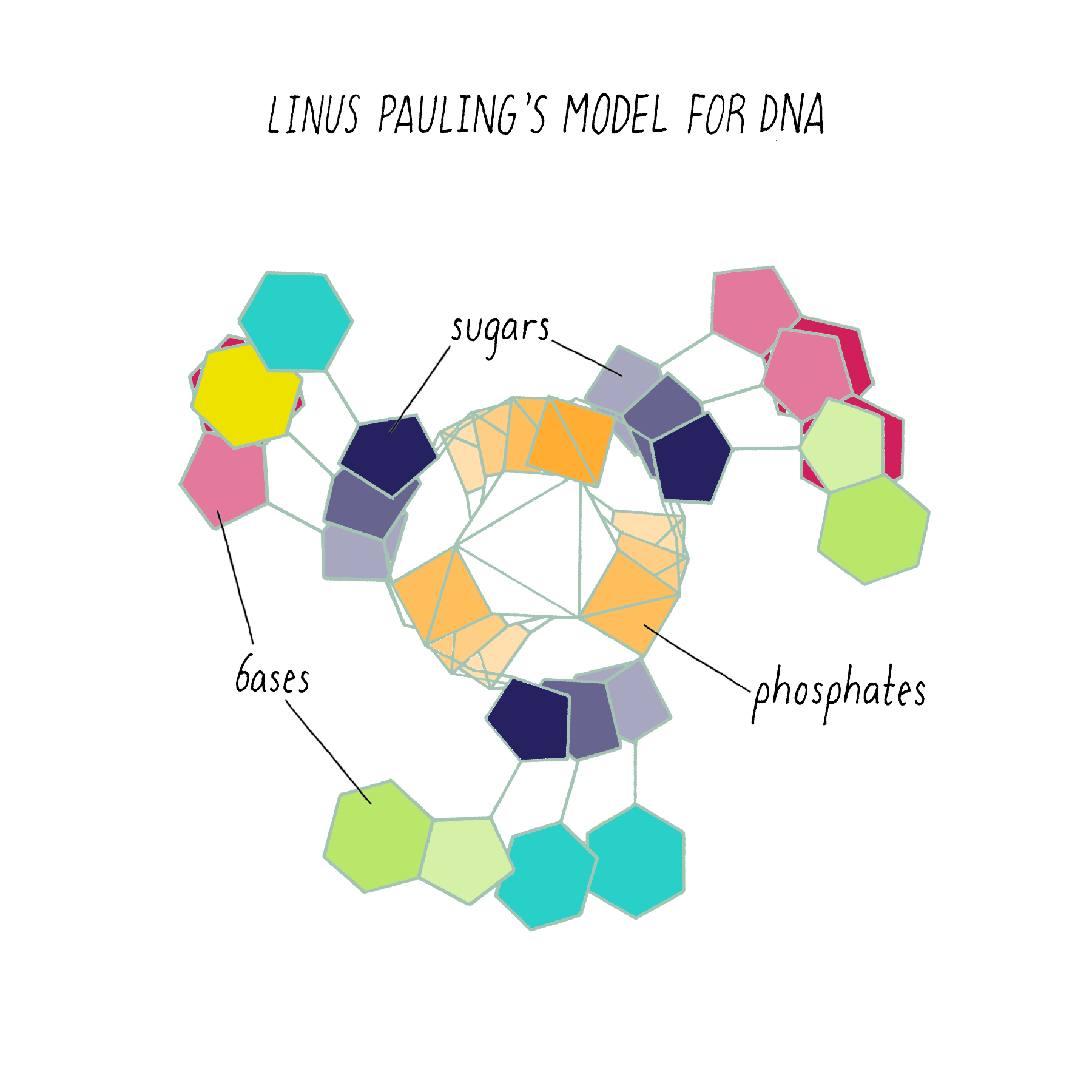
Why did Pauling, the hero of the protein alpha-helix 2 years earlier, come up with the wrong DNA structure? Pauling did not have access to the informative DNA photograph of Rosalind Franklin, which will be discussed below. But his model also was inconsistent with known chemistry, such as the attractive and repulsive forces between atoms (placing the negatively charged phosphates in the center would repel and not hold the chains together). His model also did not incorporate his own idea (described earlier) about genes having complementary halves.
Some speculate that Pauling felt that he was in a race for the DNA structure and needed to get something out fast. However, a more plausible explanation was that DNA was not a big priority for Pauling. Pauling might have thought that DNA would be another biological helix, perhaps interesting from a structural point of view, but not a structure that would unlock the "secret of life." Matt Meselson (author of the Key Experiment of How DNA Replicates) was a graduate student with Pauling at the time and was unaware that his advisor was even working on DNA (personal communication). Pauling himself later said, "we weren’t working very hard at it. I wasn’t putting much of my own time in determining the structure."; (Eighth Day of Creation; see references). If he anticipated that the DNA structure would reveal the riddle of heredity, he likely would have made solving its structure a higher priority.
The Success: The Path to the DNA Double Helix
With better X-ray equipment, Franklin and Wilkins improved the X-ray photographs of DNA over those produced by Florence Bell and William Astbury. However, their initial photographs were still difficult to interpret. Franklin then realized that the photographs were blurry because they were a mixed image produced by two different forms of DNA in the fiber. By varying the amount of humidity in a controlled manner, she then found conditions for making pure fibers of the two forms of DNA, which she called the "A-form" and the "B-form". This experimental advance by Franklin was a major contribution and an important turning point in the story.
On May 2, 1952, Franklin and her graduate student Raymond Gosling developed photograph 51, an >60-hour X-ray exposure of the DNA B-form, which revealed a pattern of short lines that formed an "X" (Figure 10). An entire theatrical play (Photograph 51) has been written on this image, one of the most famous in biology.
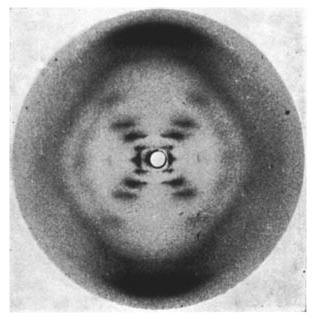
However, photograph 51 did not produce a eureka moment; its clues were not easy to spot at the time. Franklin decided to focus on the A-form of DNA, which she thought was more information-rich and might provide a direct solution to the structure. Failing to make much progress on the A-form, Franklin returned to analyzing the simpler B-form in 1953, at about the same time that Watson and Crick began their sprint to the double helix described below.
The "magical month" of the double helix began on January 30, 1953; on that day, Jim Watson took a train to King’s College to share the news of Pauling’s incorrect DNA manuscript with Wilkins and Franklin. On that visit, Wilkins showed photograph 51 to Watson, not knowing that this information was interpretable to Watson. Watson, exploding with excitement, rushed back to Cambridge to share the news of what he saw, clear evidence that DNA is a helix. Watson convinced Bragg, the Director of the Cavendish Laboratory, to allow a renewed effort at DNA modeling. They thought that Pauling would soon realize the mistakes in his DNA model and potentially figure out the correct structure. Watson and Crick were back in business modeling DNA on February 1, 1953. At that point, they knew that DNA was a helix but were potentially still a long way from a complete solution. Remarkably, four weeks later, they arrived at a successful model. On February 28, as legend has it, Crick proclaimed in the Eagle Pub that he and Watson had discovered "the secret of life." The major events in the DNA double helix story are laid out in the timeline in Figure 11.

Three significant puzzle Pieces came together to make the final model.
Puzzle Piece 1: DNA is a Double Helix
Crick:
After the first failed attempt at a DNA model, Crick returned to X-ray crystallography of proteins, in particular, keratin, the major protein of hair. Keratin is made up of helices of polypeptide chains. Crick, Cochran, and Vand developed a theory for what the X-ray pattern of a helix should look like; a defining feature is an X-shaped pattern of spots.
Watson:
Watson, a bacterial geneticist by training, neither a physicist nor a crystallographer, began learning X-ray diffraction from Crick and others at Cavendish. Thanks to Crick’s work on helical structures and through their constant discussions, Watson understood that an X-shaped pattern of spots in an X-ray photograph signified a helical structure. Primed with that knowledge, Watson instantly knew that DNA was a helix when he saw photograph 51 on January 30 at King’s College.
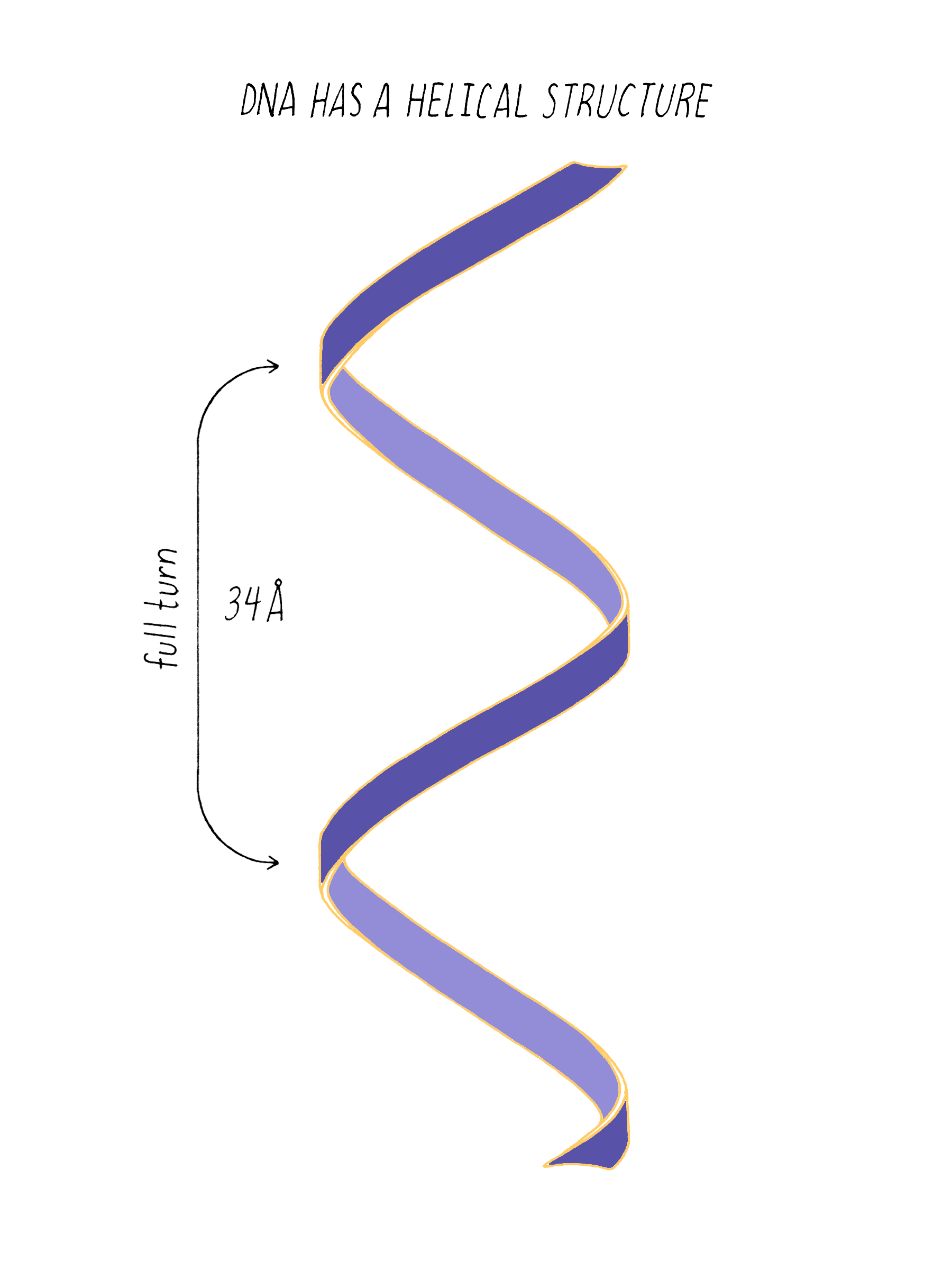
The regularly spaced lines that comprise the X-shaped pattern of photograph 51 also provided Watson with another critical piece of information: the repeat distance of the helix (34 Å; see Dig Deeper 3). This is the distance in which the DNA chain twists around and comes to the same relative point along the axis (Figure 12). Now Watson and Crick knew three critical numerical parameters for making a 3D model- the repeat distance of the helix (34 Å), the width of the molecule (20 Å), and the ladder spacing between the bases (3.4 Å).
Watson quickly returned to model building. However, based upon his inspection of photograph 51, Watson did not know whether the helical structure of DNA was made of one, two, or three chains. He guessed that it might be two chains oriented in the same direction. However, his model was not working out. Perhaps something was wrong.
Puzzle Piece 2: DNA is Composed of Two Strands That Run in Opposite Directions
Crick:
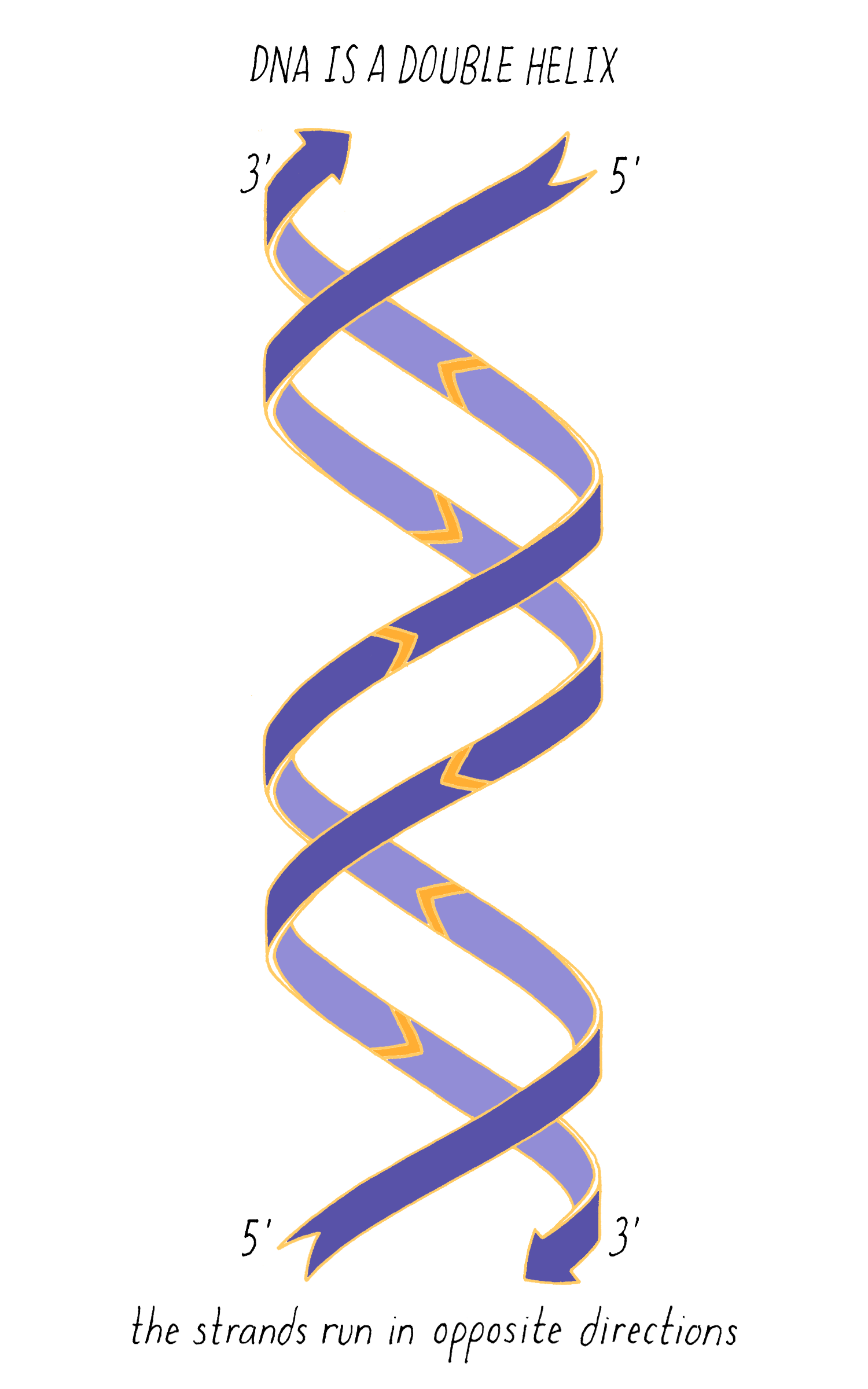
With Watson stuck in his modeling, it was Crick who provided the next critical insight. The leadership group at the Cavendish Laboratories received a progress report from King’s College which became available to Crick. The information had been previously publicly presented and at first glance might not have seemed very useful. However, Crick seized upon a phrase describing A-form DNA as "face-centered monoclinic". This technical detail provided information on the symmetry of molecules in the DNA fiber. Crick realized, which Wilkins and Franklin did not glean at the time, that this symmetry meant that DNA was composed of two identical parts, one matching the other but in a reverse orientation. Crick deduced that DNA must be made of two strands oriented in opposite directions (Figure 13). Extrapolating that the symmetry of A-form DNA also is true for B-form DNA, Watson and Crick were now confident that DNA is a double helix and now knew the orientation of the strands.
Explorer’s Question: You are building a spiral staircase model of the DNA double helix with a repeat distance of 34 Å. In a spiral staircase, the steps are offset at a specific angle. What angle would you introduce between the bases in your model of DNA? (Hint: Use information from Florence Bell on the distance between bases as well as the repeat distance revealed by photograph 51 of Rosalind Franklin).
Answer: One complete turn of the double helix is 34 Å. The distance between the bases is 3.4 Å. Therefore, ten bases make a complete turn of the helix. The angular offset between two adjacent bases would be ~36° (360° (one complete rotation)/ 10 bases).
Puzzle Piece 3: Complementary Base Pairing Holds the Two Strands Together
The X-ray data of the A- and B-form DNA revealed a double helix with a repeat distance of 34 Å and the strands running in opposite directions. But Watson and Crick lacked further details. Where are the bases? What is holding the two strands together?
Watson’s and Crick’s 1951 model and Pauling’s 1953 model both placed the phosphates at the center. Having been alerted by Franklin to the problems with this orientation, Watson and Crick now began building models with the bases placed at the center and the phosphates on the outside.
Watson began by stacking bases every 3.4 Å, as in Florence Bell’s model (Clue 3). But unlike Bell’s model, which was of a single strand, Watson had to look for plausible ways in which the bases might interact to hold the two strands of the double helix together. A well-known attractive force is a hydrogen bond, which involves a donor hydrogen interacting with an "acceptor" (usually an electronegative atom such as nitrogen (N) or oxygen (O)) (see the Knowledge Overview). Watson began to look for possible ways in which hydrogen bonds could be formed between bases across the double helix.
To start this effort, Watson looked up the chemical structures of bases from a well-regarded textbook. (As you will see soon, you cannot always trust what you read in a textbook!). Watson came up with what he thought was a solution: like interacts with like. In this homo (meaning "same") base pairing idea, adenine pairs with adenine, guanine with guanine, cytosine with cytosine, and thymine with thymine (Figure 14).
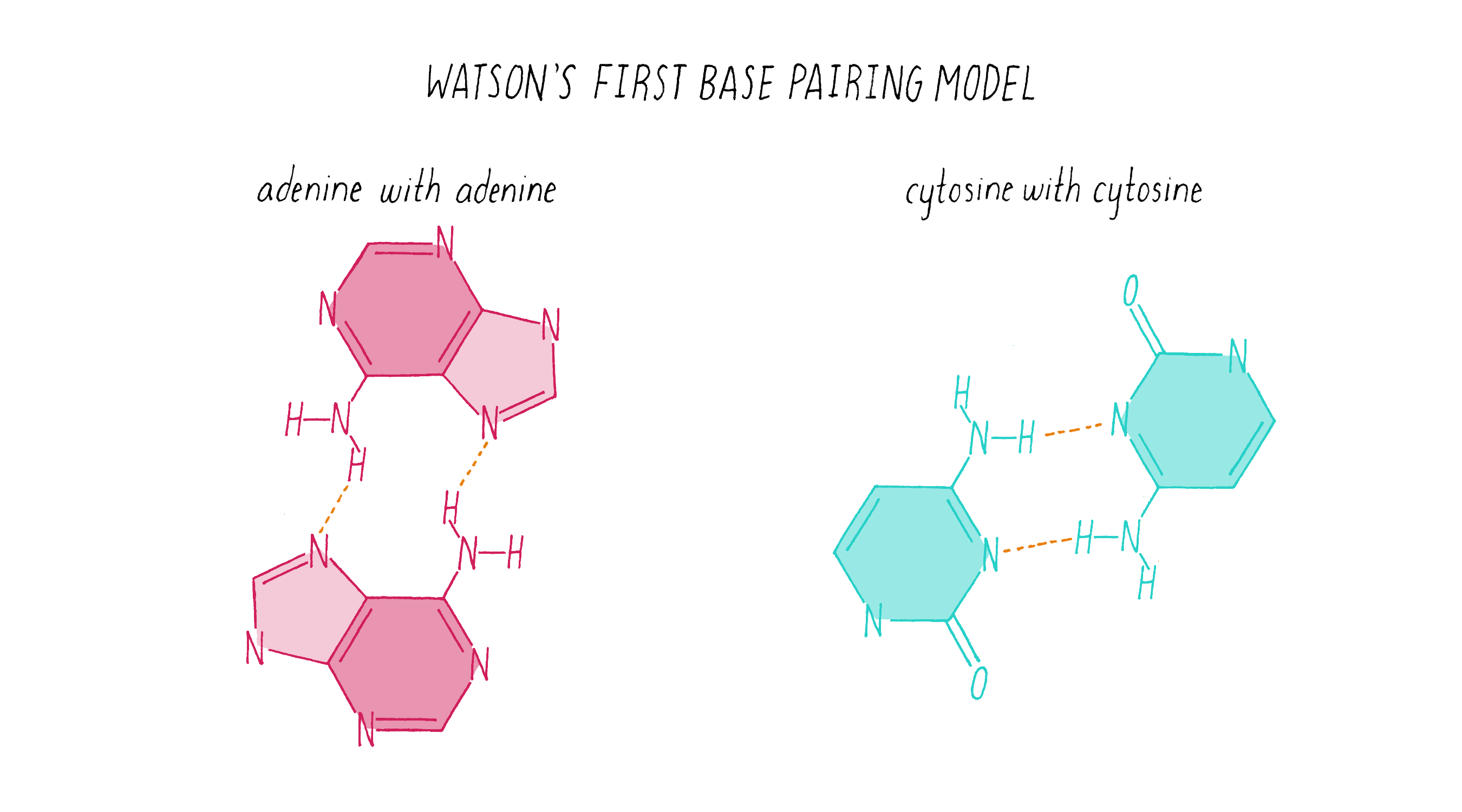
Explorer’s Question: Does the homo base-pairing model of DNA provide a solution for how a gene might be replicated?
Answer: Yes. The homo pairing of bases is a form of complementarity and provides a simple rule for how a new strand of DNA could be copied from a preexisting strand. In this model, the two strands would be composed of identical sequences of letters.
Explorer’s Question: Can you think of a problem with this homo base-pairing model?
Answer: The model has a major problem for creating a double helix of a constant diameter. The purine pairings (G-G and A-A) are wider (since they are double rings) than the pyrimidine pairings (T-T and C-C) (single ring bases), resulting in a variation in the width of the molecule.
Explorer’s Question: Is the homo base-pairing model consistent with Chargaff’s data?
Answer: No. The homo base-pairing model does not provide a rationale for why the ratio of G to C and A to T are both equal to 1.
Watson was excited about the homo base-pairing model because it provided a simple rule for replicating a gene. However, there were problems with the model, as discussed in the Explorer’s Questions above.
Then, another fortuitous event in the magical month transpired. Jerry Donohue, an expert in the chemistry of nucleotide bases, was visiting the Cavendish Laboratory and assigned to work in the same room as Watson and Crick. When Watson presented his homo base-pairing model, Donohue told him that the "textbook" chemical structures for guanine and thymine were likely incorrect. Nucleotide bases can adopt two slightly different chemical structures, called tautomers, which contain the same set of atoms but differ in the position of a single hydrogen atom. While this difference is small, it matters a lot for figuring out the right set of hydrogen bonds between the bases. Donohue said that the guanine and thymine tautomers used in Watson’s homo base-pairing model were likely incorrect. If one has the wrong idea or model, the best thing that can happen is to discard it quickly. Watson’s model unraveled within a span of 2 days.
Now using the correct base structures described by Donohue, Watson began building new models on February 28, 1953. He began playing with pieces of cardboard cut out in the shapes of the four bases, looking for ways in which they could interact. Then came what can be considered the "eureka moment" in the DNA double helix story, as re-enacted by Jim Watson many years later in this short video (Video 3). On that very same day, Watson and Crick enjoyed a celebratory pint at the Eagle Pub, announcing the "secret of life" to a beer-drinking clientele who may not have fully appreciated the historical significance of the moment.
What Watson discovered on February 28, 1953, were some very simple base-pairing rules. Guanine (the tautomeric structure pointed out by Donohue) formed matching hydrogen bonds with cytosine. Using the correct structure for thymine, Watson also saw that adenine and thymine formed a perfect hydrogen bond pair. Watson and Crick realized that the G-C and A-T hydrogen bonding was the key ingredient needed to hold the double helix together (Figure 15).
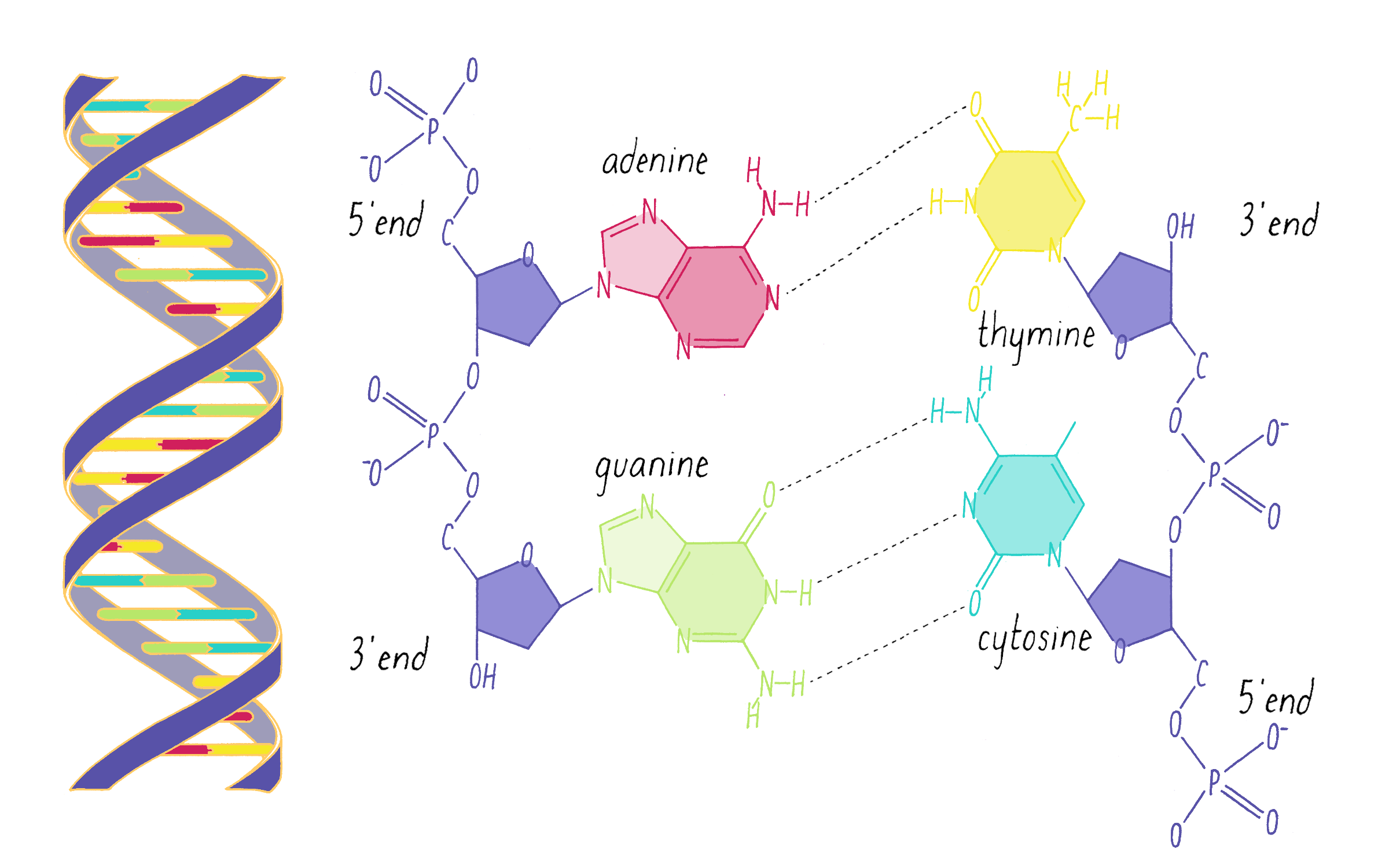
Explorer’s Question: Did the new G-C and A-T base-pairing model provide a solution for how a gene might be replicated?
Answer: Yes. Like the homo-pairing model, this provides a simple rule for creating a newly replicated strand from a template strand. If the parental strand has a G, then the replicating partner strand needs to place a C opposite to it.
Explorer’s Question: Why was the A-T and G-C pairing model a better structural solution than the homo-pairing model?
Answer: The two purine-pyrimidine pairs (G-C and A-T) are identical in width. No matter what the order of nucleotides in the DNA, the width of the double helix will be constant.
Explorer’s Question: Is the A-T and G-C base pairing model consistent with Chargaff’s data?
Answer: Yes. In this model, there is a structural explanation of why the ratio of G to C and A to T is equal to 1. However, Watson and Crick did not initially incorporate the Chargaff data in making the model, as Crick explains in The Eighth Day of Creation: "The paradox of the whole thing is that when it came to build, we did not initially use the idea. We didn’t until we were driven towards it. … And I can remember the moment when we realized that, by God, we could build a structure, in which the bases would be complementary. And explain Chargaff’s one-to-one ratios that way."
Today, scientists use computers to build molecular models. However, in the days before computers, Watson and Crick built a physical model, much like building a model airplane. The machine shop at the Cavendish Laboratory cut tin plates into the shapes of bases, true to their relative dimensions but ∼300,000,000-fold larger than life. Watson and Crick attached the bases onto a backbone of metal sugars and phosphates, creating a twisting sculpture. It was a model of the atomic biological world conceived in the human mind. The metal double helix was beautiful, but its shape was not the important breakthrough. Rather, it was the powerful idea of complementarity (the A-T and G-C pairing). Pauling, and others, thought that complementarity might provide a mechanism for copying a gene. Now, in the Watson and Crick model, complementarity provided a physical explanation for how the two strands in the DNA double helix were held together. In addition, it provided a possible explanation for how genes replicate, which will be discussed in the next sections What Happened Next and the Knowledge Overview. Bell, Astbury, Watson, Crick, Pauling, Wilkins, and Franklin could not envision at the outset of their studies that a clear understanding of gene replication would emerge from the structure of DNA. However, the fact that a simple explanation did emerge made the double helix model all the more believable. As Watson said,
"There had been far too many days when Francis and I worried that the DNA structure might turn out to be superficially very dull, suggesting nothing about either its replication or its function in controlling cell biochemistry. But now, to my delight and amazement, the answer was turning out to be profoundly interesting." In The Double Helix (see references).
Many readers of this Narrative are probably familiar with the game "Clue." In this game, players accumulate clues, until one player feels that s/he has accumulated enough clues to announce a solution to the crime: "it was Mr. Green who committed the murder with a Rope in the Library." After making the announcement, the player anxiously waits to see if their solution can be disproved by any of the other players.
Watson and Crick announced their first model of a DNA structure in 1951. It was too early; they had not accumulated enough clues, and they were out of the game, at least temporarily. Now, Watson and Crick were ready to announce their new model, and see if anyone could shoot it down: DNA is a double helix with G-C and A-T base pairs in the center. They presented it to Donohue and the nucleotide expert Alexander Todd (see Clue 1), both of whom could not find a flaw from a chemistry point of view. They called in Lawrence Bragg, the head of the Cavendish Laboratory, who could offer no objection from a structural point of view. It was then time to invite Wilkins and Franklin to Cambridge to see their model. Franklin and Wilkins knew instantly that the Watson and Crick model was consistent with their data and thus was likely, although not certainly, right.
What Happened Next?
A manuscript on the double helix model was submitted by Watson and Crick to Nature. A coin toss decided the order of the authors on the paper. (We could be calling it the Crick and Watson model had the coin landed differently.) Franklin and Wilkins prepared separate papers on the X-ray data, which were submitted to Nature at a similar time. All three papers were published together on April 25, 1953.
The Watson and Crick paper on the model DNA structure essentially occupied a single printed page (see Guided Paper). It left the reader with this provocative message:
"It has not escaped our notice that the specific pairing that we have postulated immediately suggests a possible copying mechanism for the genetic material."
Watson and Crick refrained from proposing more detailed thoughts on replication in their first paper, because they had not yet seen all of the X-ray data by Wilkins and Franklin. They were still somewhat uncertain if their model was correct. However, once they saw the Wilkins and Franklin papers, they became more confident. Watson and Crick then published their more provocative ideas on DNA replication in a subsequent Nature paper published on May 30. In this paper, Watson and Crick provided more information on the base pairing. However, they postulated only two hydrogen bonds for the G-C base pair, which Pauling later correctly pointed out should be three. Even classic papers have imperfections. In their replication model, Watson and Crick suggested that the hydrogen bonds holding the chains together are broken (somehow) and that each individual strand serves as a template to create a new complementary strand. This model was later called "semi-conservative replication" (Figure 16).
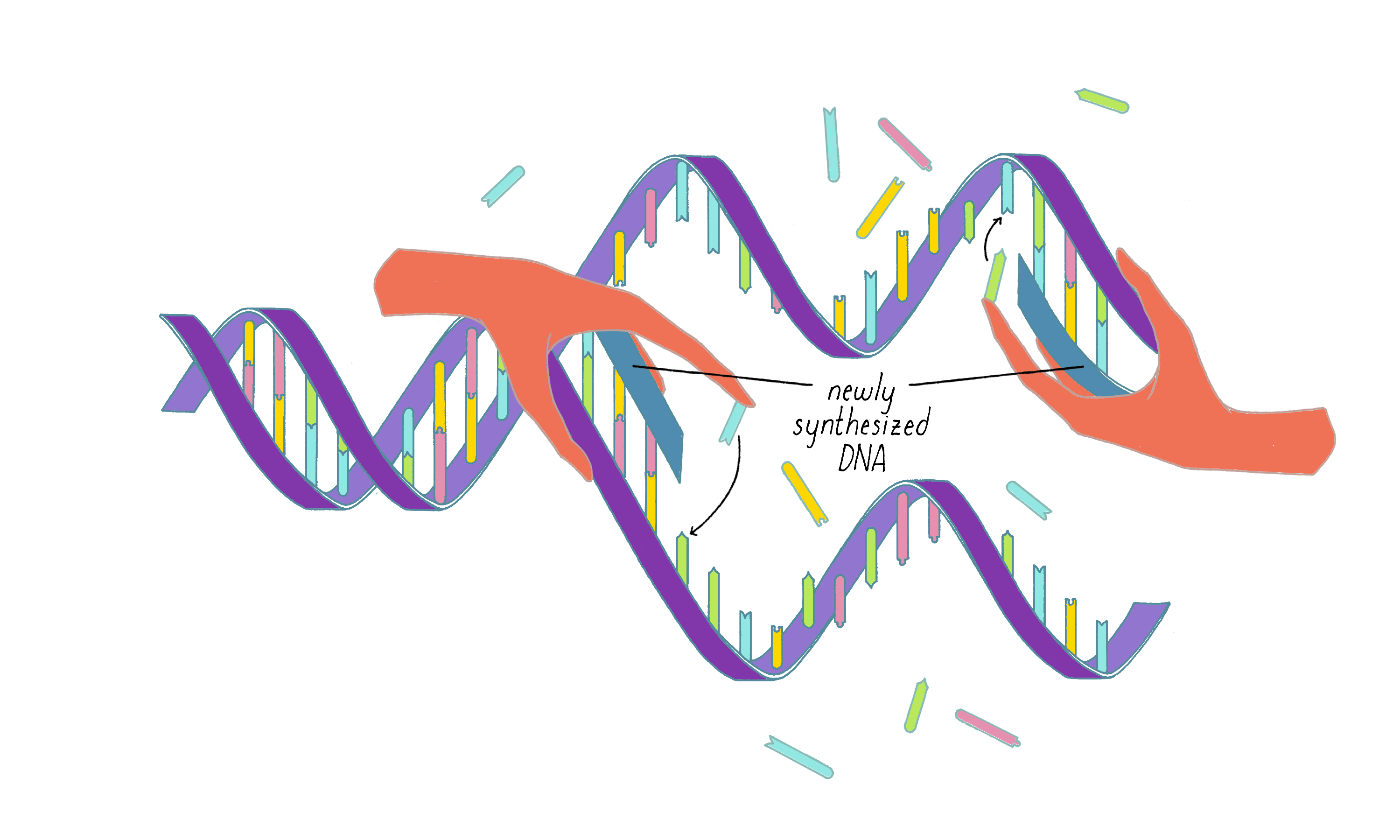
The model of the DNA helix and its replication were received with a mixture of enthusiasm and caution. Pauling gracefully acknowledged that the Watson and Crick model, and not his, was correct. Other scientists remained skeptical, realizing that data from the X-ray work could not fully support the details of the Watson–Crick model. Moreover, the Watson–Crick hypothesis for DNA replication lacked any supporting evidence.
Models need to be tested with experiments. Two 24-year-old scientists, Matthew Meselson and Frank Stahl, rose to the challenge. Their experiment (coined "the most beautiful experiment in biology") supported the Watson and Crick hypothesis of semi-conservative replication and ruled out competing models (Video 4). You can learn about this experiment first hand from Meselson and Stahl in their Key Experiment in XBio. The 1958 Meselson and Stahl publication had a significant impact on the field. After seeing these clear results, skepticism about the Watson and Crick model waned, and the acceptance of the double helix model ushered in a new era of molecular biology. In the same year as the Meselson–Stahl experiment, Arthur Kornberg announced the discovery of an enzyme called DNA polymerase that can copy DNA (discussed in the Knowledge Overview).
Interestingly, the Watson–Crick model was only definitively proven in 1980, when the atomic structure of the B-form DNA was determined using high-resolution X-ray data from a crystal rather than a fiber (shown later in Video 6). It was the result that Rosalind Franklin dreamt of, but it would only happen 30 years later.
What happened to the main characters after the DNA double helix? Watson and Crick remained friends but parted ways for different scientific pursuits. Watson began to work on the structure of RNA. But lightning did not strike twice. RNA has a complex and variable structure, and his efforts did not succeed. However, Watson made other important contributions, including co-discovering messenger RNA. Francis Crick graduated with a PhD in 1954 at the age of 37 and next turned his attention to how DNA serves as a code. Crick proposed the Central Dogma—which states that the information of DNA flows unidirectional from DNA to RNA to protein (although the later discovery of reverse transcription proved an exception to this rule). The code is briefly discussed in the Knowledge Overview but will be covered in more depth in another Narrative.
Franklin and Wilkins also parted ways. Because of her difficulties at King’s College, Franklin made arrangements to join Birkbeck College in June 1952, only a month after obtaining photograph 51, and then moved in mid-March, 1953, just after Watson and Crick developed their DNA model. As part of this transition, Franklin was asked to summarize her work on January 28, 1953, and then to transfer her DNA data to King’s College. After leaving King’s, Franklin left DNA behind. She switched to studying the structure of viruses, publishing 17 papers and doing groundbreaking work in this field. Wilkins continued to work on and make important contributions on DNA, including developing more convincing evidence for the double helix using X-ray data and more precise model building.
Franklin died of cancer at the age of 37 in 1958, 4 years before the Nobel Prize was awarded for DNA structure. Before her death, there was no bitterness between Franklin and Watson and Crick; on the contrary, she developed a friendship with both, and even stayed at Crick’s home in Cambridge while recovering from her treatments for cancer.
The regular and simple structure of DNA was much easier to solve than those of proteins and RNAs, which have more complex shapes. However, relatively soon after the double helix, Max Perutz and John Kendrew at the Cavendish Laboratory managed to solve the 3D structure of myoglobin, an oxygen-carrying protein from muscle that is similar to hemoglobin. Now, over 150,000 protein structures have been solved, most experimentally; predicting protein structure is still very difficult even with modern supercomputers. Each one of those protein structures tells an interesting story, although DNA still occupies the throne as the most illuminating and impactful structure of all time.
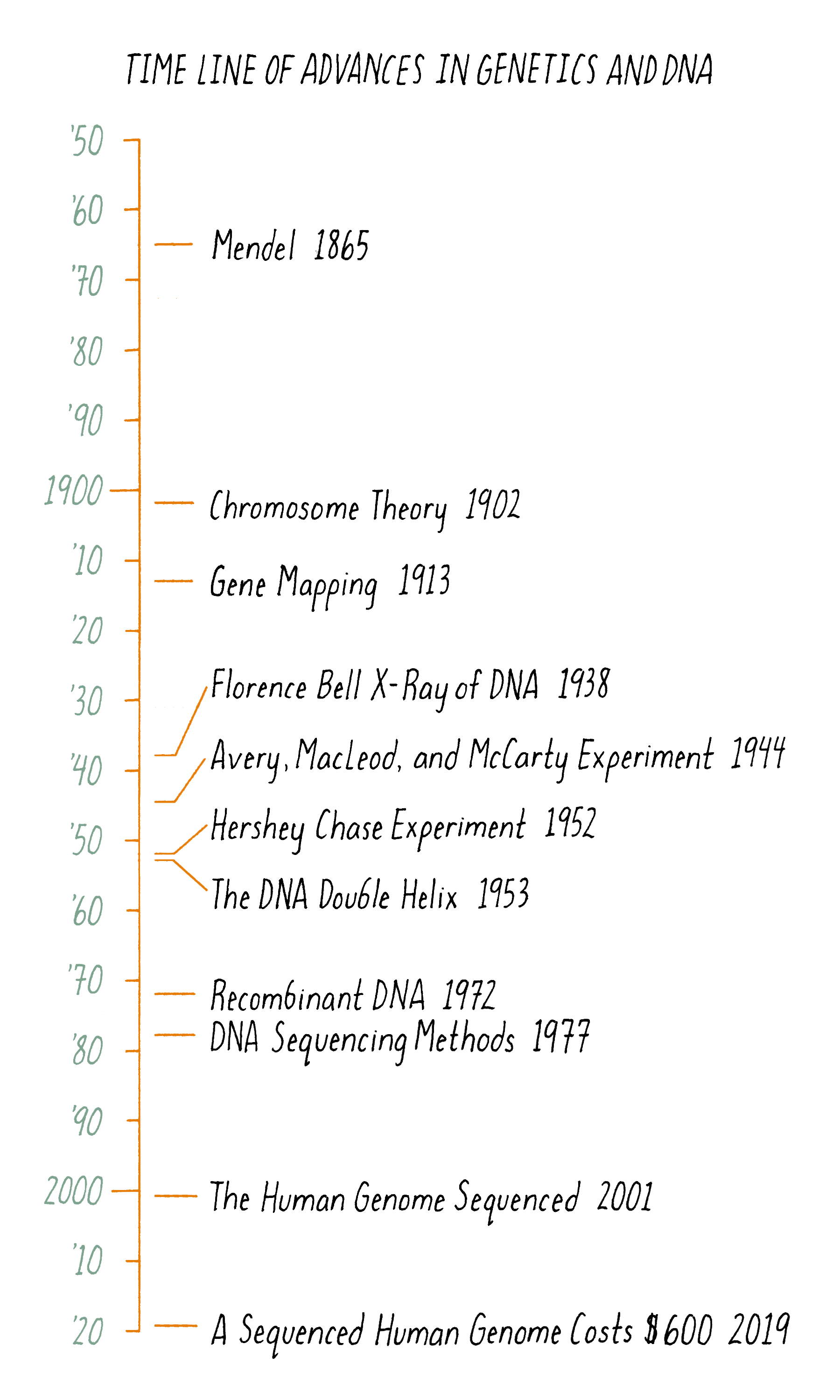
Figure 17 summarizes the scientific journey of DNA, beginning with Mendel’s peas. But the timeline begins at least a couple of billion years earlier. At that time, a DNA molecule existed with the same structure as found today. That DNA propagated and evolved, eventually producing beings capable of deciphering the structure of the molecule responsible for their very existence.
Part II: Knowledge Overview —
The structure and replication of DNA
Deoxyribonucleic acid (DNA) is a polymer of nucleotides. DNA in prokaryotes and eukaryotes consists of two polymer strands that twist around in a double helix (see Journey to Discovery). Some viral genomes, however, are made of single-stranded DNA. Most of the information that you need to know about the structure of DNA is contained in this excellent ∼5 min video made by MITx (Video 5) and also will be covered in text form below.
What does DNA do?
DNA performs three major functions in biology.
1) DNA is an information storage device. In this role, DNA must be very stable so that it can store information for the lifetime of an organism (which in the case of some trees could be a few thousand years).
2) The information in DNA instructs the production of other biological molecules. Specifically, RNAs are copied from DNA in a process called transcription. The copied information in many RNAs is used to instruct the production of proteins.
3) DNA can make copies of itself, with the help of specialized proteins, in a process called DNA replication. When one cell divides into two, DNA replication allows each cell to possess a complete copy of the genome.
The Features of the DNA double helix
The building blocks of DNA are nucleotides
A nucleotide is composed of three component parts:
1) A nitrogenous base, of which there are four varieties in DNA (Figure 18). Adenine and guanine, the double-ring bases, are called purines, and cytosine and thymine, single-ring bases, are called pyrimidines. The double bonds in the bases make them planar (2-dimensional).
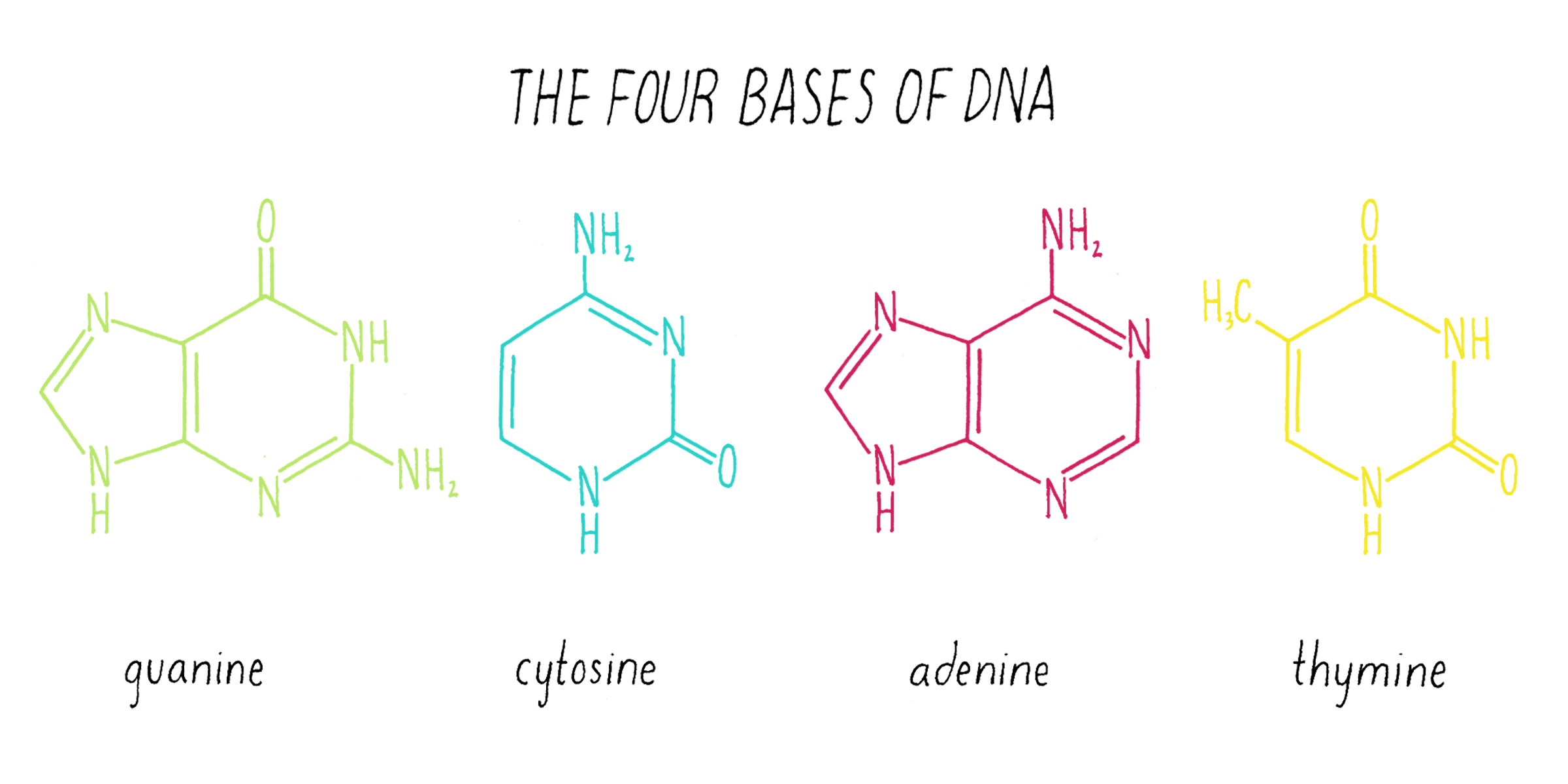
2) A base is connected to a deoxyribose, which is a five-membered sugar ring (Figure 19). The carbons in the ring are labeled with a number and a prime symbol (′). Deoxyribose lacks a hydroxyl (OH) at the 2′ position. Ribonucleic acids (RNA) are similar to DNA but have a hydroxyl (OH) group at the 2′ position. The combination of a base with a sugar is called a nucleoside.
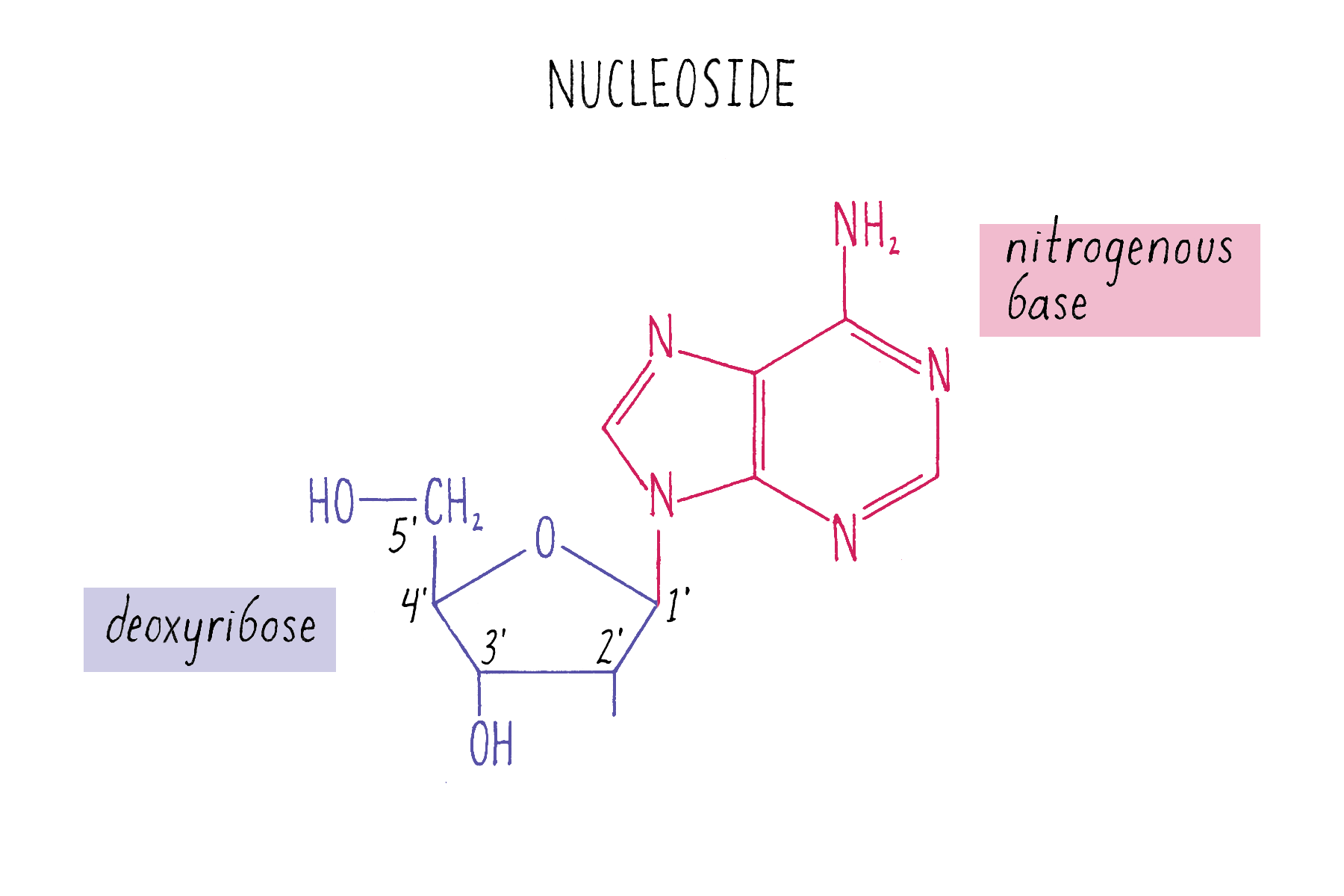
3) A nucleotide has a phosphate joined to the 5´ carbon of the sugar (Figure 20). Nucleotides are the basic building blocks of DNA. The coupling of a single phosphate is called a nucleotide monophosphate; two phosphates, a nucleotide diphosphate; and three phosphates, a nucleotide triphosphate. Later, we will encounter adenosine triphosphate in a role as an energy source that drives DNA replication.
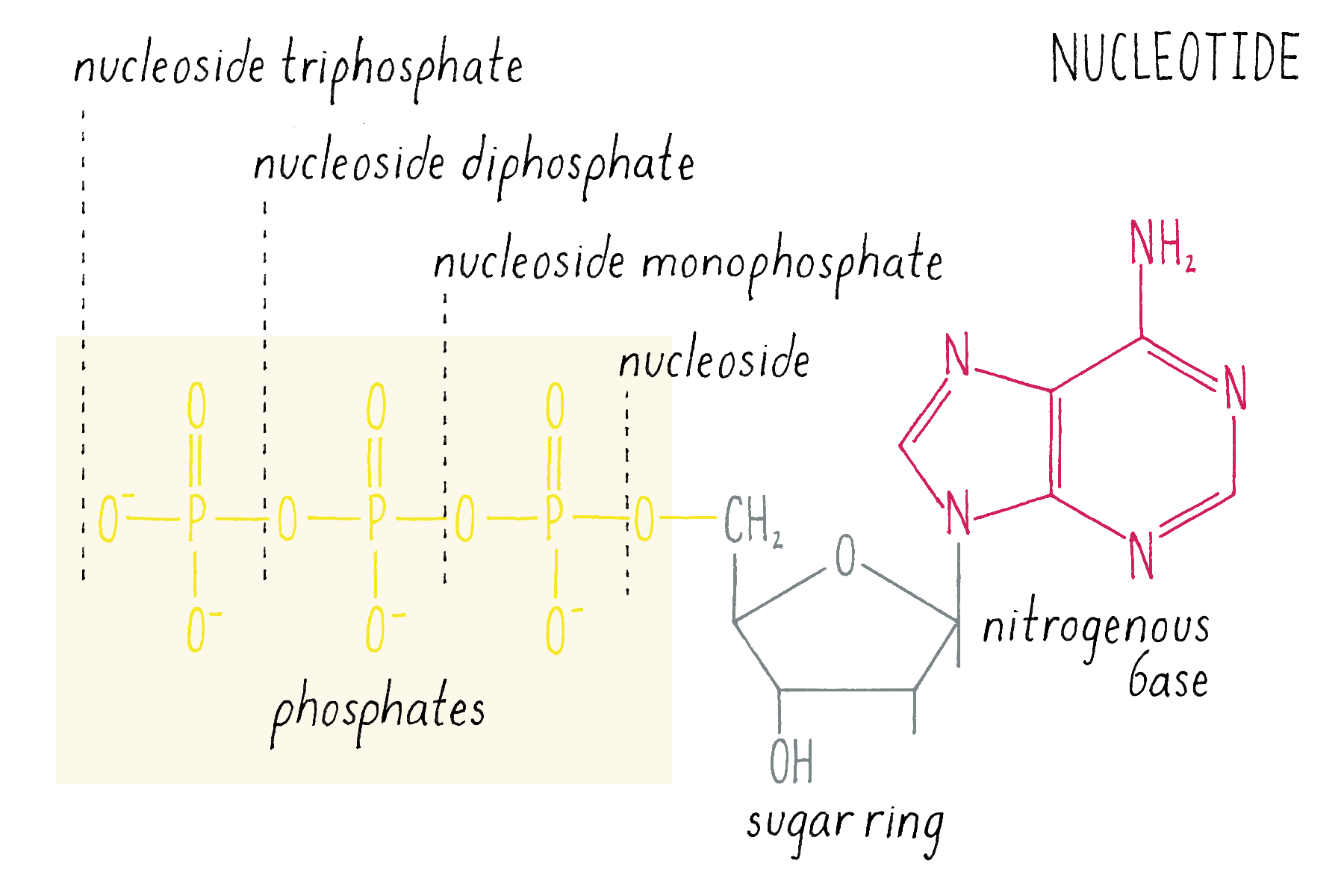
DNA is a polymer of connected nucleotides
Each human chromosome is a very long DNA double helix with many connected nucleotides. The largest human chromosome is made of 250 million nucleotides in each of its two strands. The phosphate group links adjacent nucleotides, connecting the 5´ carbon of one deoxyribose to the 3´ end of the next deoxyribose (Figure 21). A bond between a phosphate and a sugar is called a phosphate ester bond. Since one phosphate is bonded to two sugars, the linkage is referred to as a phosphodiester bond. The sugars and phosphates constitute the continuous backbone of the DNA molecule. Because DNA is a long chain of connected nucleotides, it also is referred to as a polynucleotide.
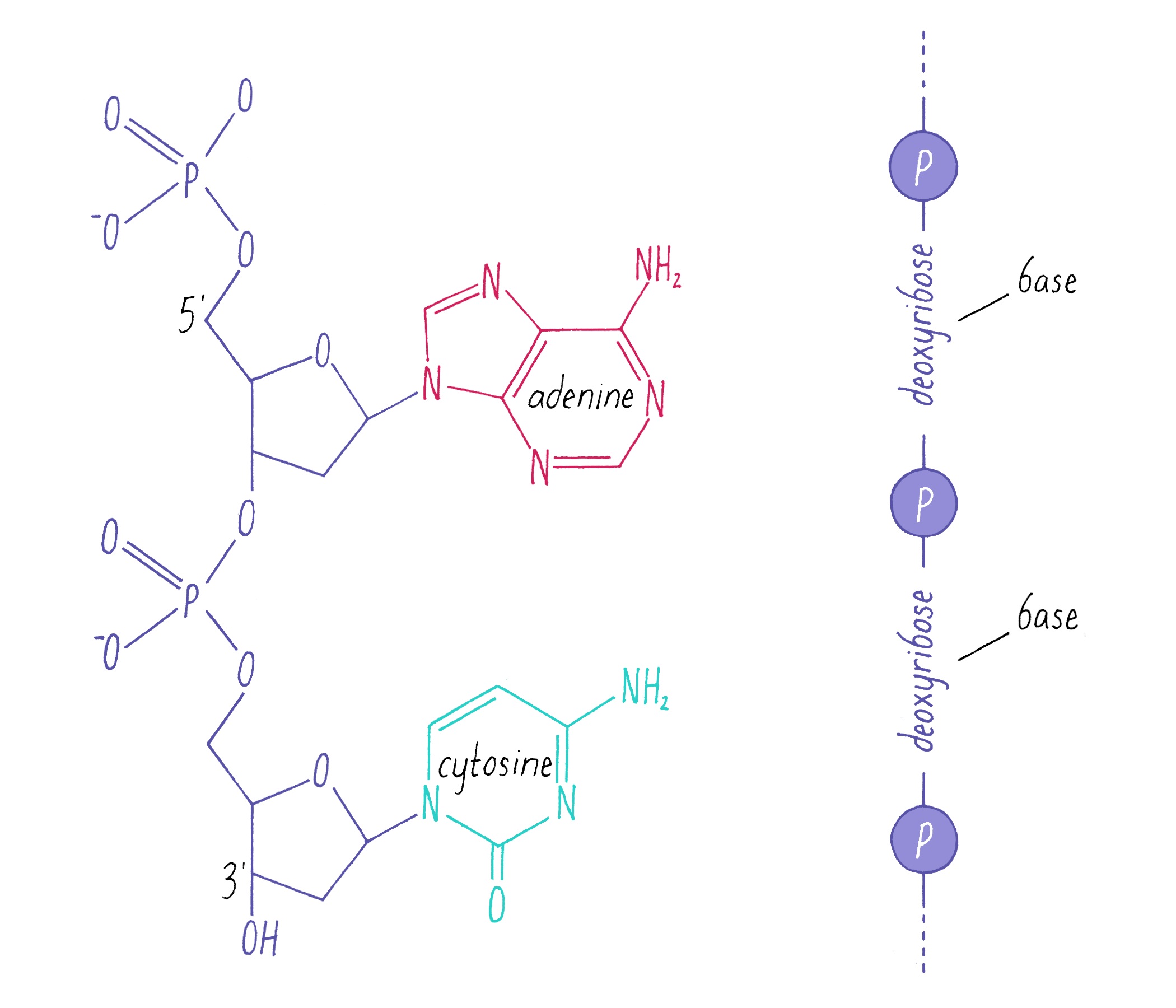
Because the phosphates are joined to the sugars through different carbon positions, there is a polarity (directionality) to the DNA chain. As seen in the chain in Figure 21, each deoxyribose has its 5´ hydroxyl (OH) pointing up and its 3´ hydroxyl pointing down.
The two strands come together to make a double helix
In biology, DNA almost always exists as a duplex known as B-form DNA (Rosalind Franklin’s terminology; see Journey to Discovery ). There are two alternative forms of double helical DNA called A-DNA and Z-DNA (Dig Deeper 4). Here we describe the basic properties of the B-form DNA double helix, which include:
• A right-handed twist of the helix. With your thumb pointing upward, the fingers in your right hand will naturally follow the twist of the double helix (Figure 22). Another way to think about the handedness of a helix is from the viewpoint of looking down its axis. If the backbone curls clockwise, the helix is right-handed. If it turns counter-clockwise, it is left-handed.
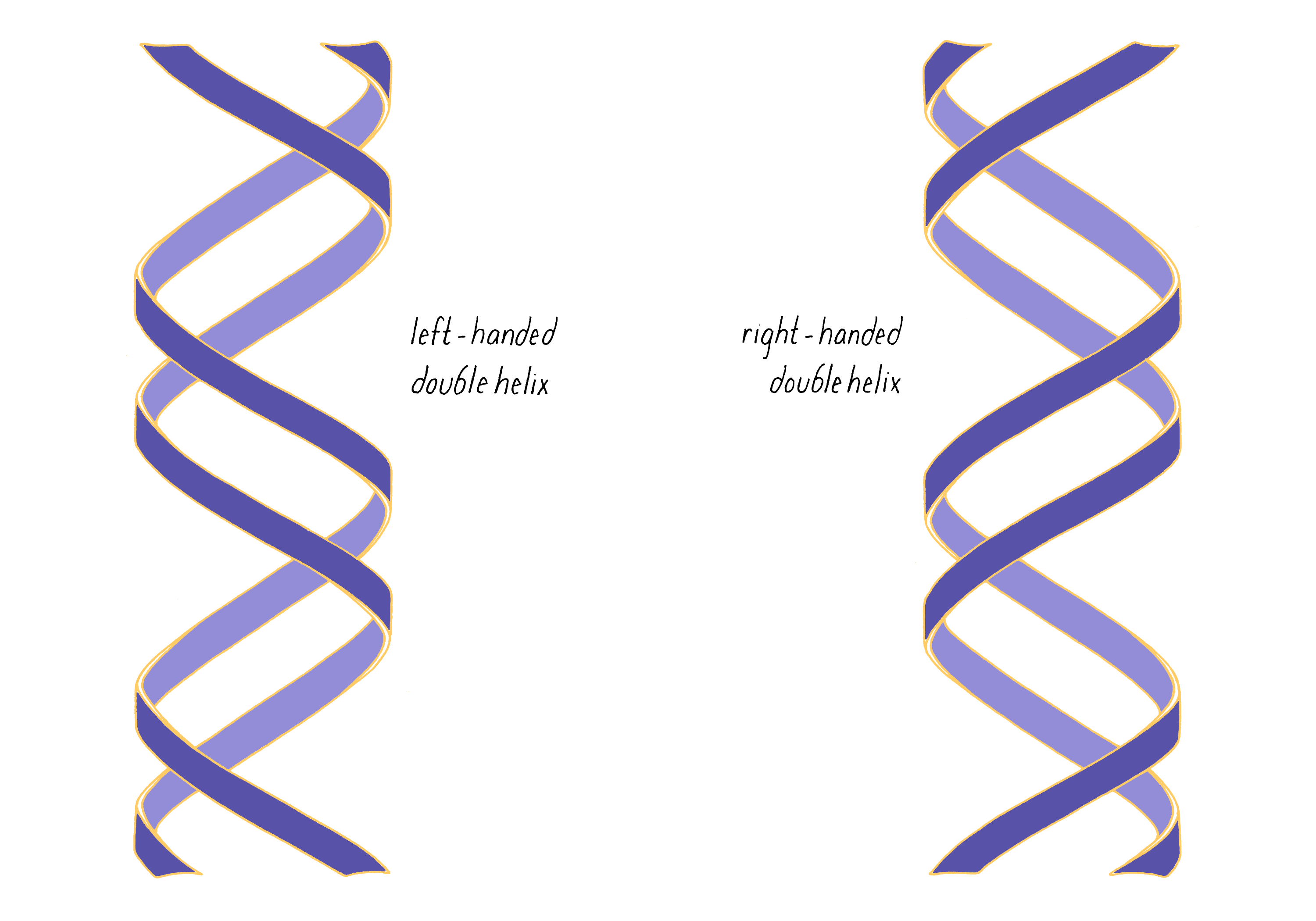
• The two DNA strands run antiparallel to one another (polarity is defined by the 5´ and 3´ orientation of the sugars in the chain, as discussed above). The helix repeats its twist approximately every 10 base pairs (Figure 23).
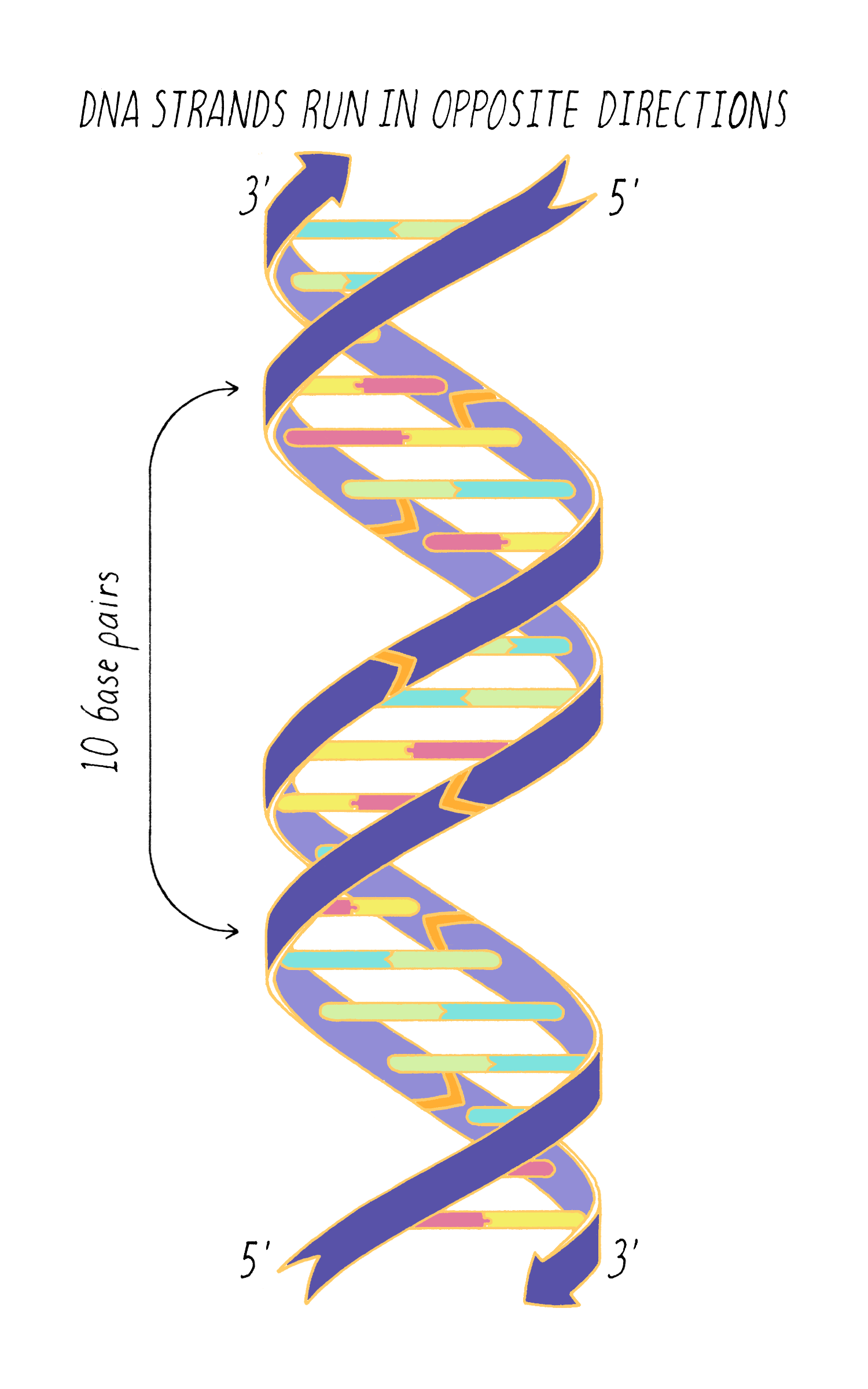
• The helix has two groves, a major and a minor groove (Figure 24), which can serve as binding regions for proteins. The bases are more accessible in the major groove. Transcription factors, which control the production of RNAs, often recognize a defined sequence of bases.
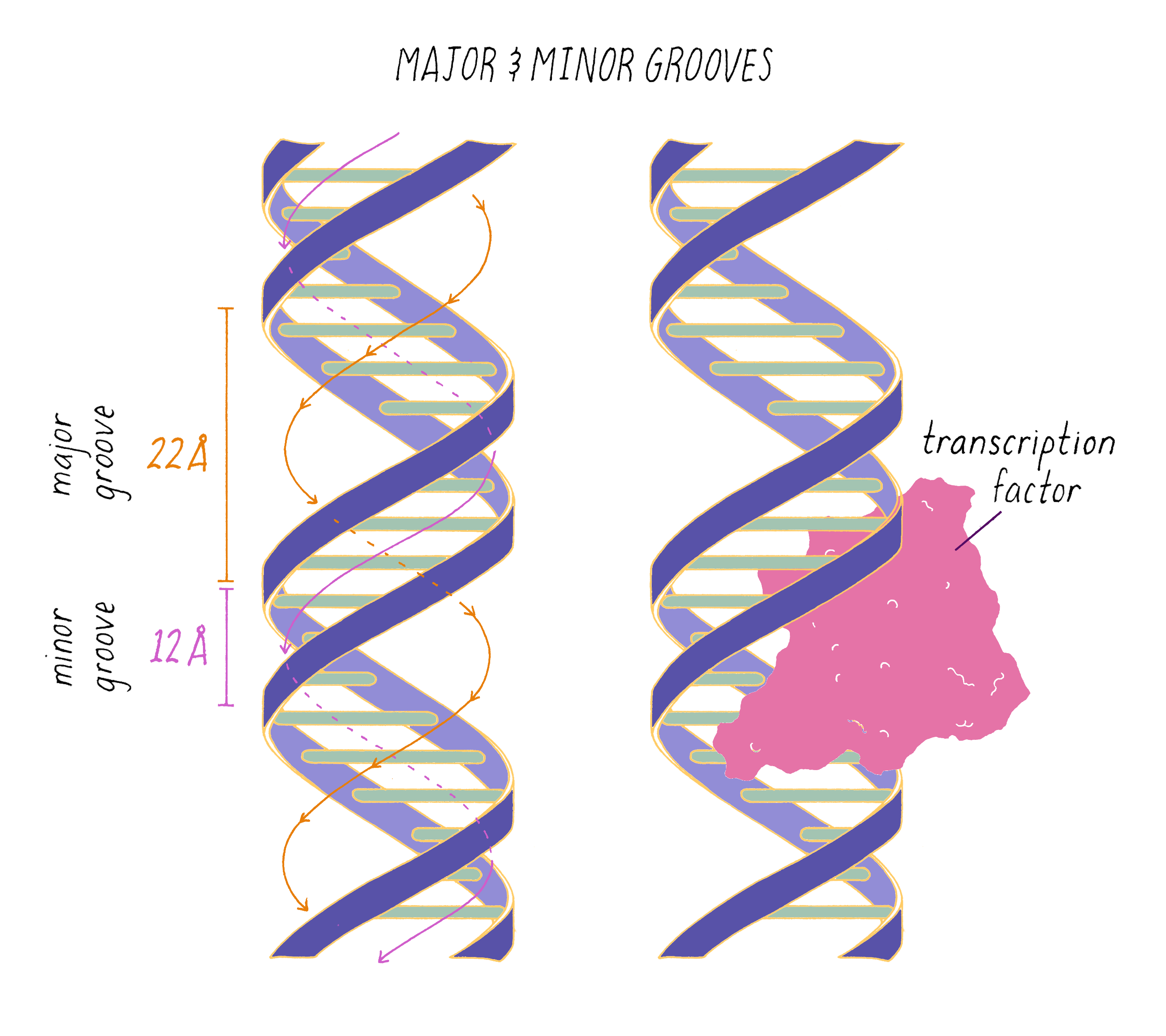
Two DNA strands are held together by hydrogen bonds and stacking interactions
The two strands are brought together by non-covalent hydrogen bonds between the bases. A hydrogen bond involves two atoms in close proximity: (1) a hydrogen atom "donor," which is covalently bound to an electronegative atom, such as nitrogen in a base, and (2) an electronegative acceptor with unpaired electrons, such as an oxygen (O) or a nitrogen (N) in a base. In Figure 25, covalent bonds are indicated by solid lines and the hydrogen bonds are indicated by dashed lines. In DNA, hydrogen bonds connect guanine with cytosine (three H-bonds) and adenine with thymine (two H-bonds) (Figure 25). Different organisms can have different G-C versus A-T content in their DNA. The higher the G-C content, the more stable the double helix will be, for example at higher temperatures.
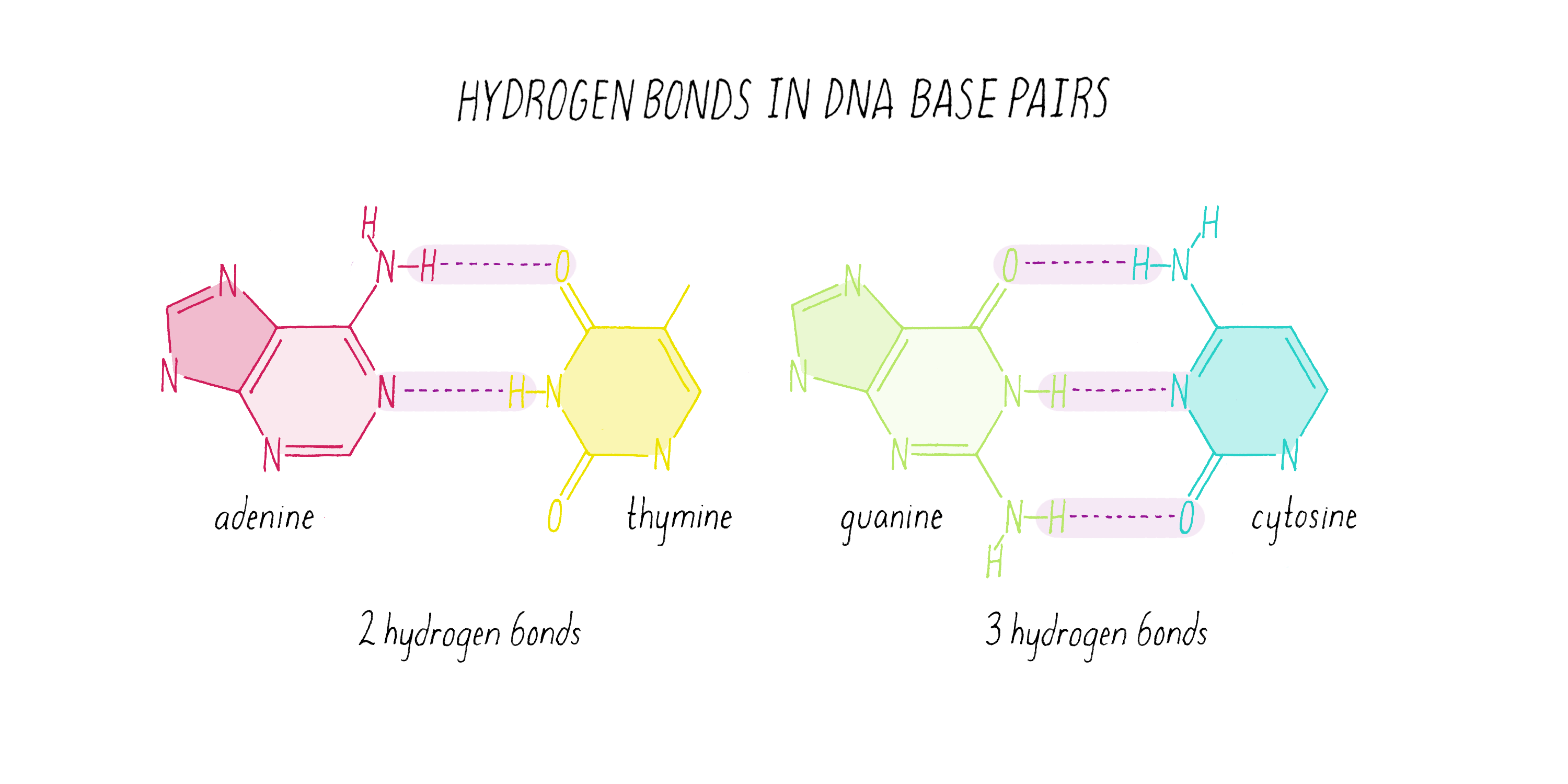
Vertical stacking interactions between the bases also help to stabilize the DNA double helix. In a "stick" representation of nucleotide structures, it looks as though there is a lot of empty space between the bases (Video 6). However, the same structure viewed in a space-filling model, where spheres reflect the extent of the electron orbitals, shows the bases are almost touching one another (Video 6). The close contacts along the long axis, called "Pi stacking" interactions, generate attractive forces that stabilize the double helix. Certain anti-cancer drugs with planar ring structures (such as doxorubicin) can squeeze in and pry apart the bases of DNA, which interferes with the replication of DNA in dividing cells.
The replication of DNA
Watson and Crick first realized that complementary base pairing of G with C and A with T provides a mechanism for the replication of DNA. Because of these base-pairing rules, each strand can serve as a template for the creation of a complementary strand.
Explorer’s Question: If the sequence of one DNA strand is known, then the sequence of its complementary strand can be easily determined. What is the complementary sequence for this strand (Figure 26A)?
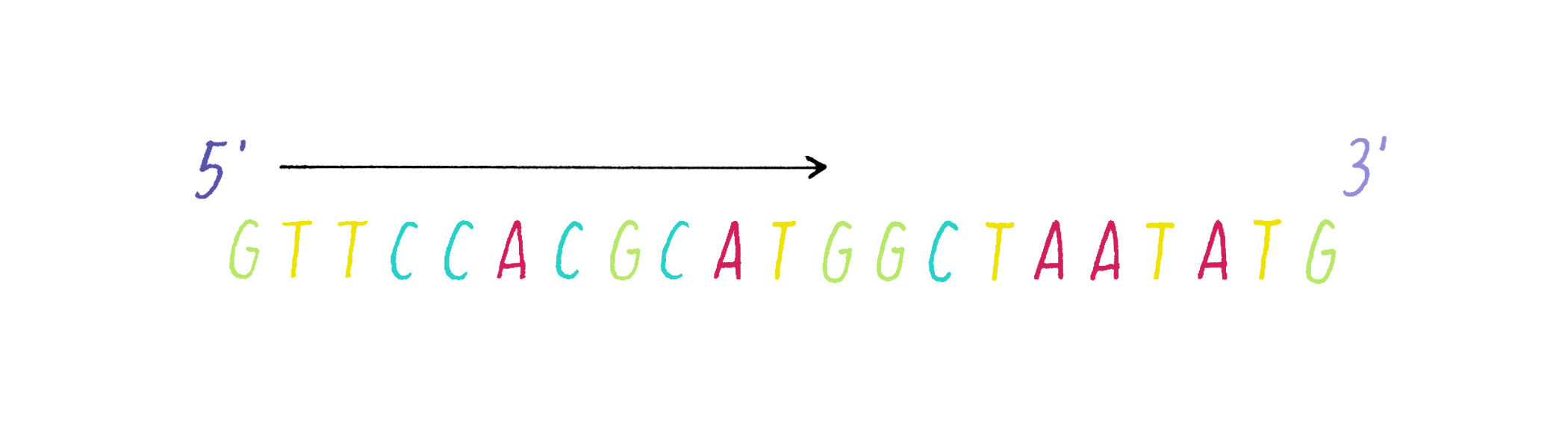
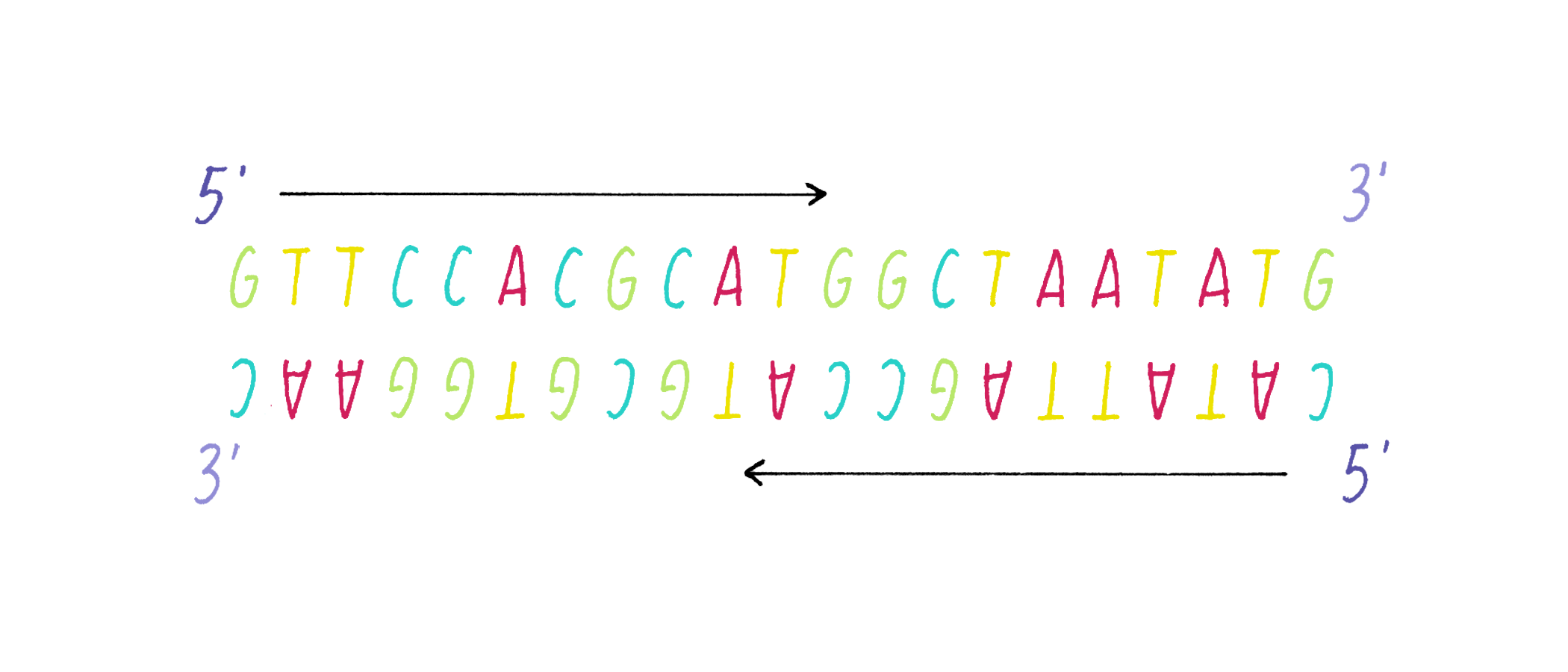
The process of DNA replication in cells requires that the two strands separate so that new strands can be copied from the parental templates. This is called semi-conservative replication (see the Key Experiment by Meselson and Stahl; Figure 27).
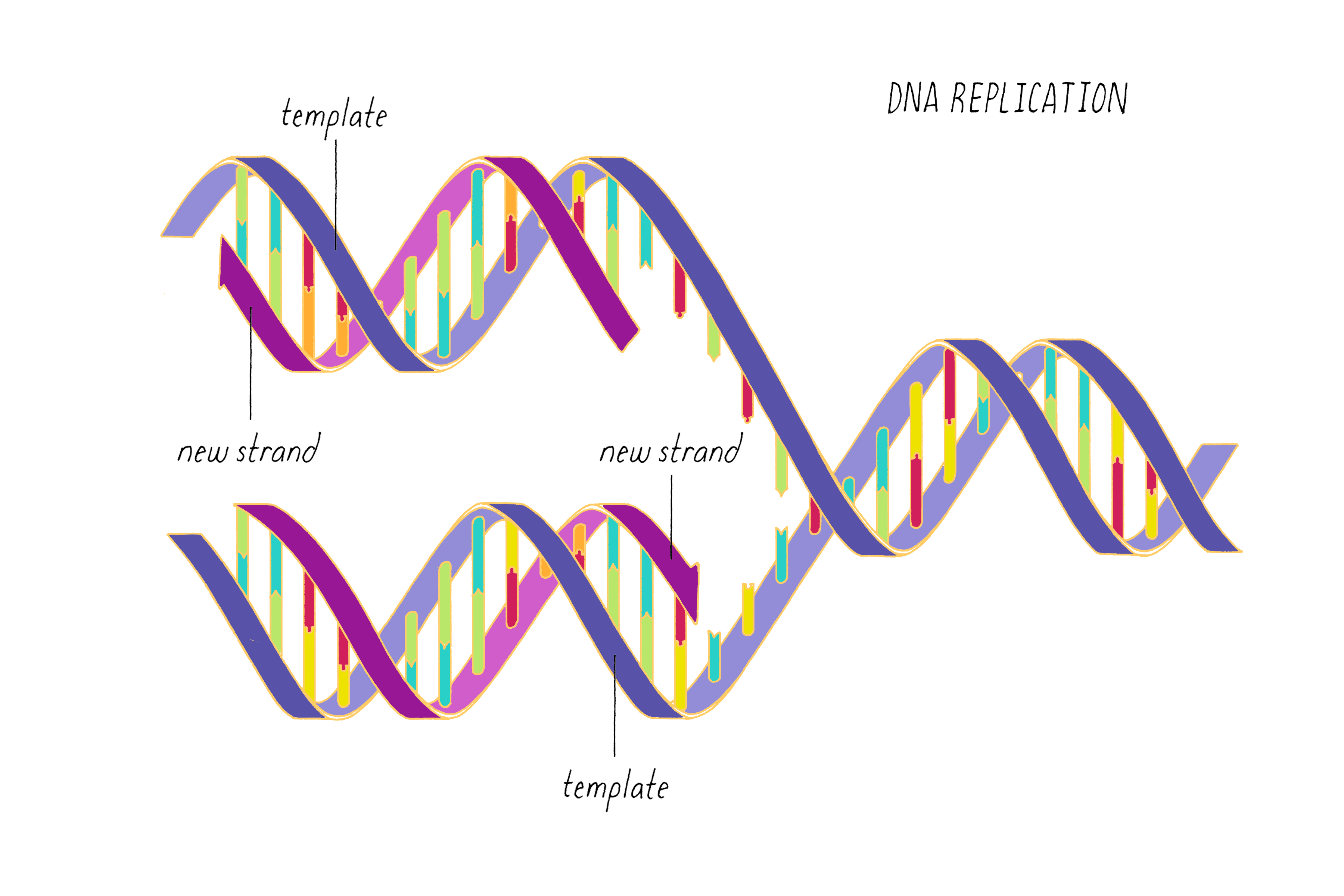
We now know a great deal about the mechanism of DNA replication, which involves many steps and many protein machines. Here we will provide a very brief overview of DNA replication; many details are left out and can be found in other resources.
Separating the two strands
As discussed earlier, hydrogen bonds and pi-stacking interactions hold two DNA strands together and energy is needed to break them apart. The energy required to break these non-covalent bonds can be provided by heat (approaching boiling temperatures), as is used in polymerase chain reaction, a method for copying DNA in the laboratory. However, cells use enzymes called helicases to separate DNA strands using energy provided by adenosine triphosphate (ATP), a nucleotide that also serves as a chemical energy source. The DNA double helix also has to untwist during replication, an action performed by another enzyme called topoisomerase. The helicase and topoisomerase enzymes reside at a junction called the replication fork, where the double helix is separated into two individual strands (Figure 28). The separated single strands are kept apart by single-stranded DNA binding proteins (not shown), which prevent the separated strands from snapping back together to reform a double helix.
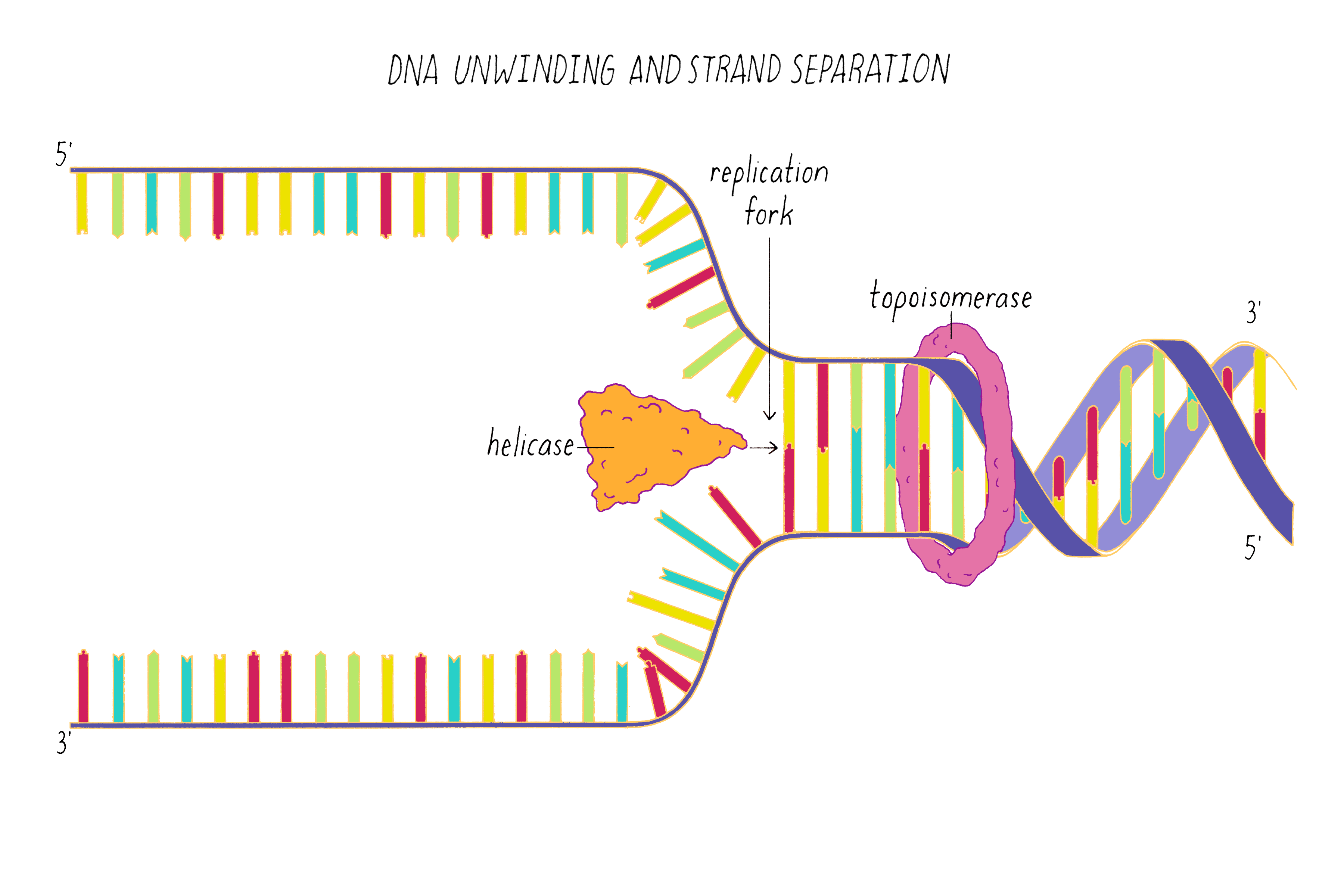
Copying the "leading" strand
Like a snowplow, the replication fork bulldozes in one direction along the double-stranded DNA, leaving two single strands of DNA in its wake.
DNA is copied by an enzyme at the fork called DNA polymerase. DNA polymerase synthesizes a new strand using the parental template to guide the insertion of each new nucleotide. The new nucleotide is added to the free 3´ end of the replicating strand (Figure 29). Thus, the new strand grows in the 5´ to 3´ direction.
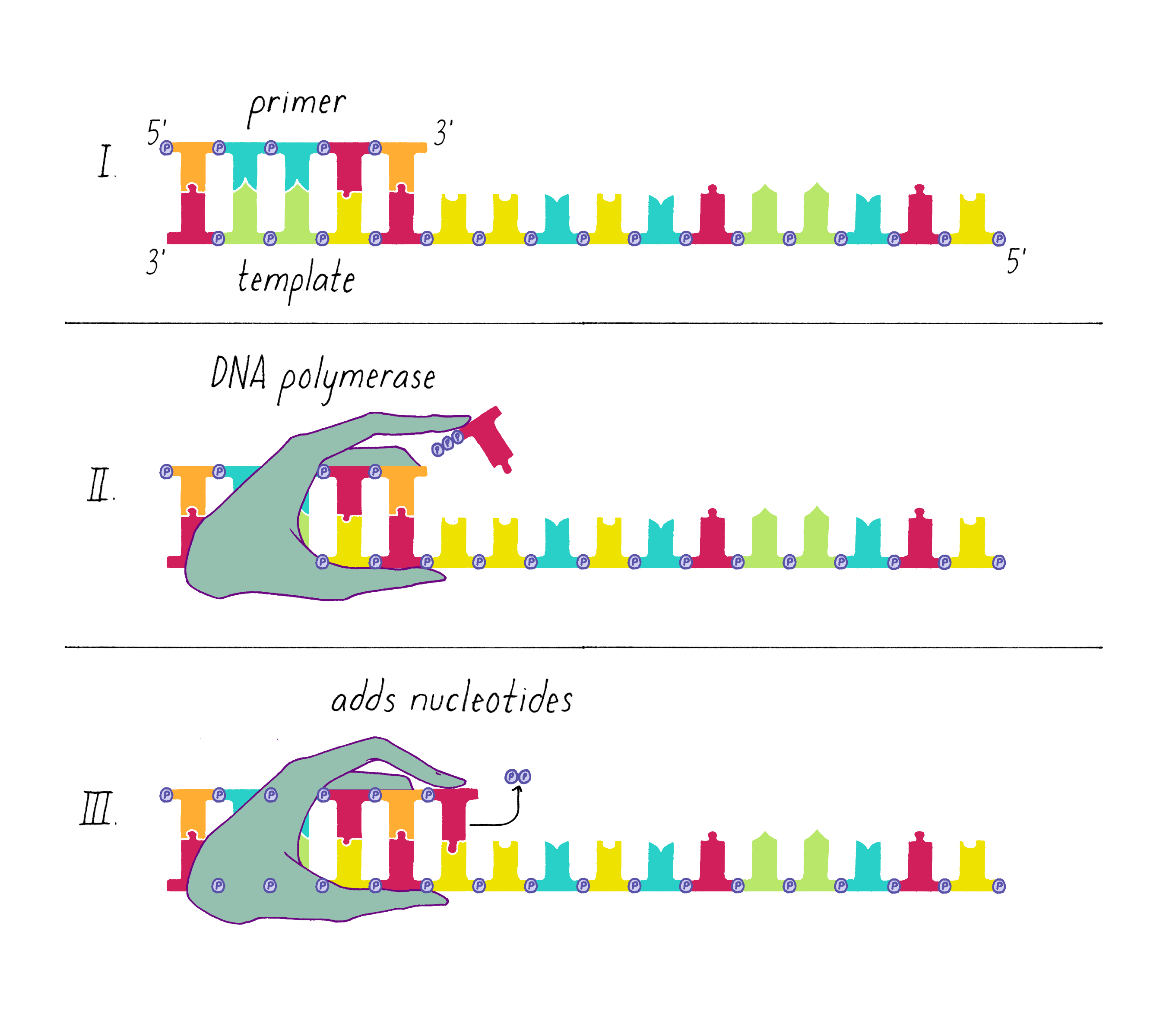
One strand, called the leading strand, has its 3´ end pointing in the direction in which the replication machinery is moving. DNA polymerase cannot start with just single-stranded DNA, and instead needs to start with a short duplex that provides the free 3´ end. A short duplex is first created by the enzyme DNA primase, which copies a short stretch of complementary RNA onto the single-stranded DNA. This RNA-DNA duplex, called a "primer," provides the initial substrate for DNA polymerase. Then, DNA copying of the leading strand proceeds continuously, since the newly synthesized DNA strand has its 3´ end oriented toward the replication fork. DNA polymerase at the replication fork can copy DNA as fast as the single strands are produced (Figure 30).
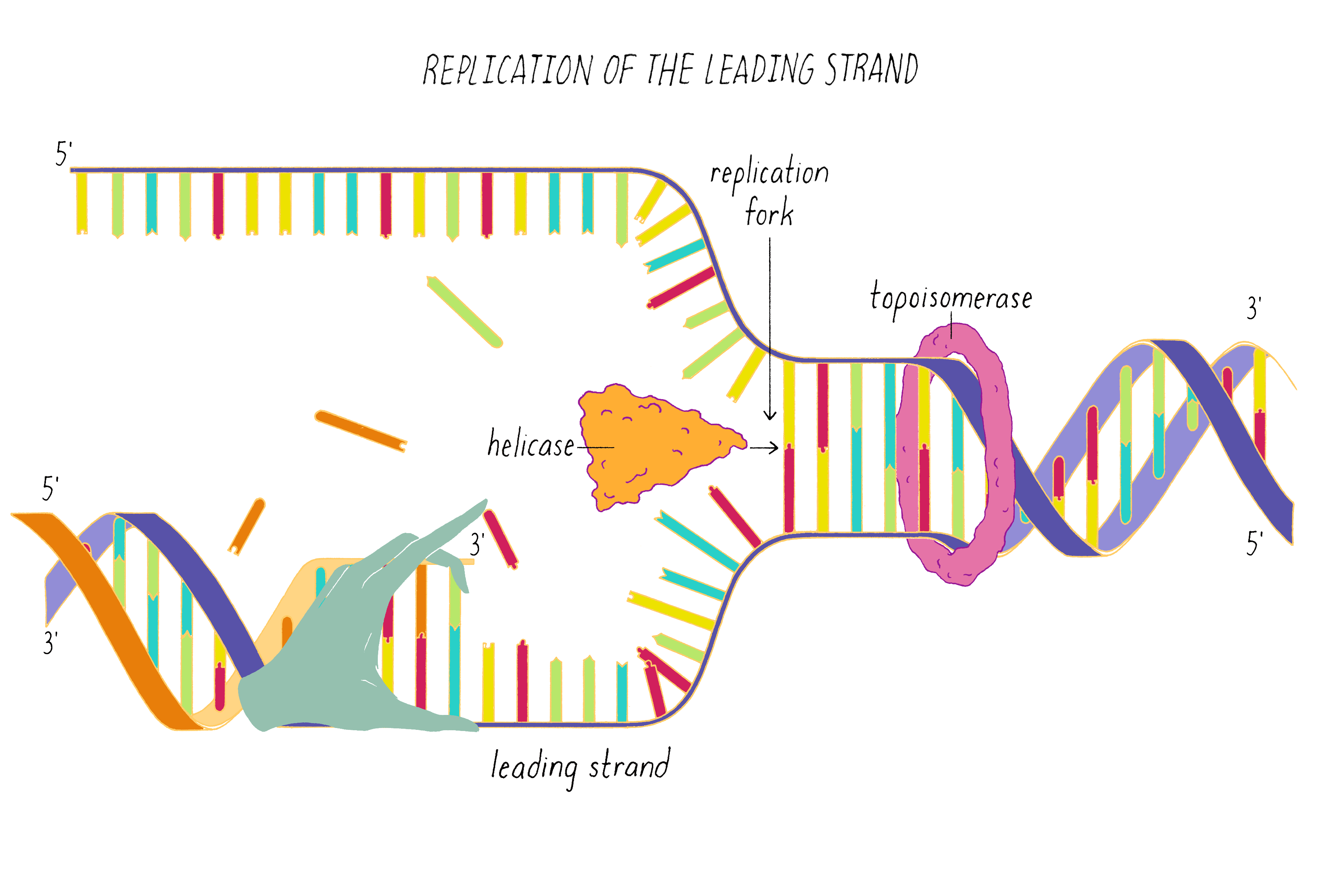
Copying the "lagging" strand
The lagging strand is replicated by a more complicated process, since the 3´ end of the newly synthesized strand is pointing opposite to the direction in which the replication fork is moving. This necessitates a discontinuous replication process. Once the lagging strand is produced by the helicase, multiple, short RNA primers are copied onto it by DNA primase. These RNA-DNA duplexes provide 3´ ends for DNA polymerase to add nucleotides (the DNA polymerase moving in the opposite direction to the replication fork; Figure 31). These discontinuous pieces of replicated DNA are called Okazaki fragments (the name of the husband-wife team who discovered them). The replication process for the lagging strand produces DNA and RNA/DNA duplexes. Next, the RNA primers are removed, and the resultant gaps are filled in by DNA polymerase.
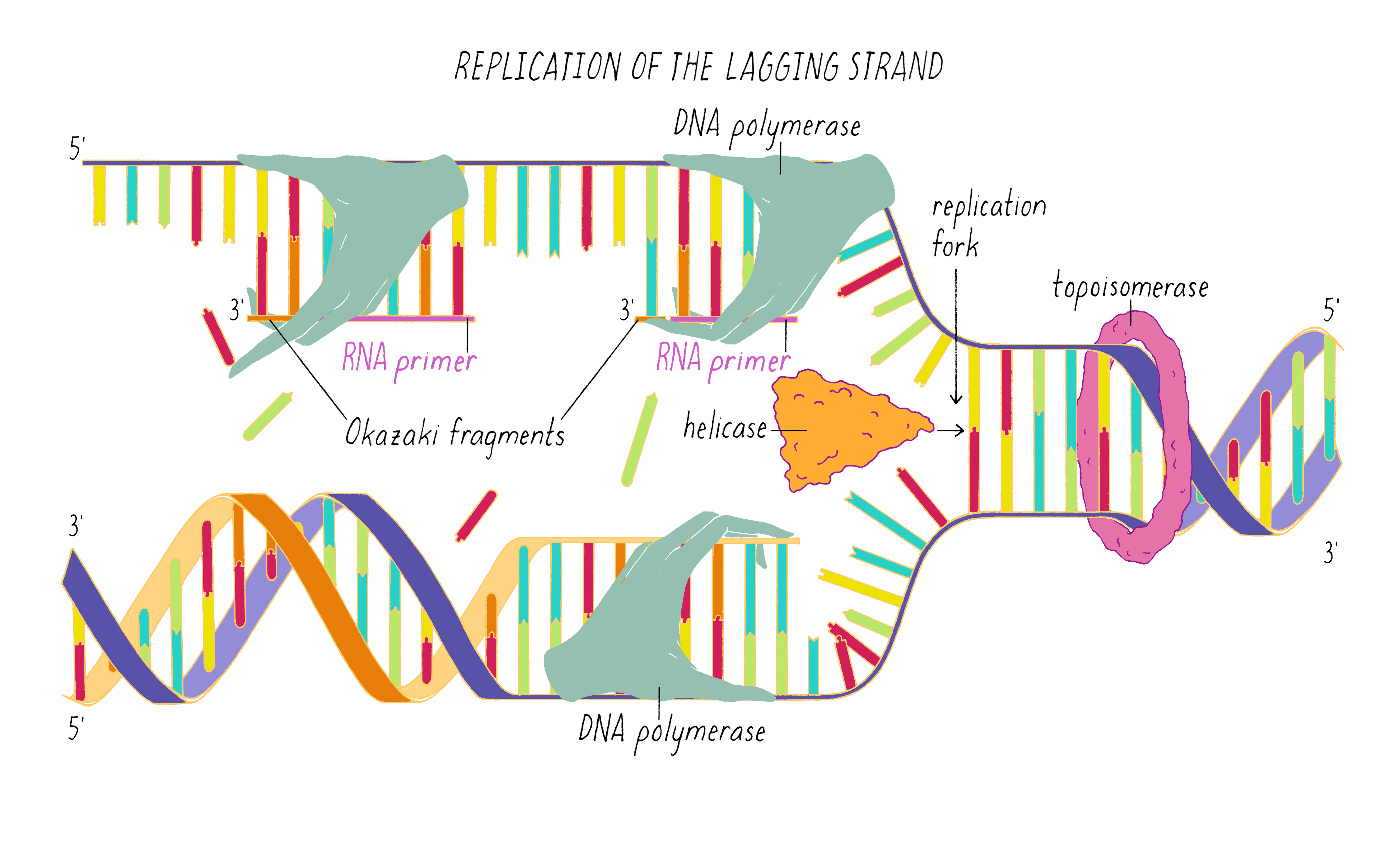
Even after the RNA is removed and DNA has been synthesized all along the lagging strand, there is a remaining problem of discontinuities between the Okazaki fragments, which needs to be corrected. DNA ligase now finishes the job of making the DNA duplex continuous. This enzyme creates a covalent phosphodiester bond between the 5´ end of one Okazaki fragment and the 3´ end of the adjacent Okazaki fragment (Figure 32). It seems as though it would be easier to invent another kind of DNA polymerase that could add nucleotides to a free 5´ end, which would allow the lagging strand to be replicated like the leading strand. However, who is one to argue with billions of years of evolution! The existing mechanism may have emerged in its present form to synthesize a new strand as well as correct errors in nucleotide addition (error correction is an important process which is not discussed here).
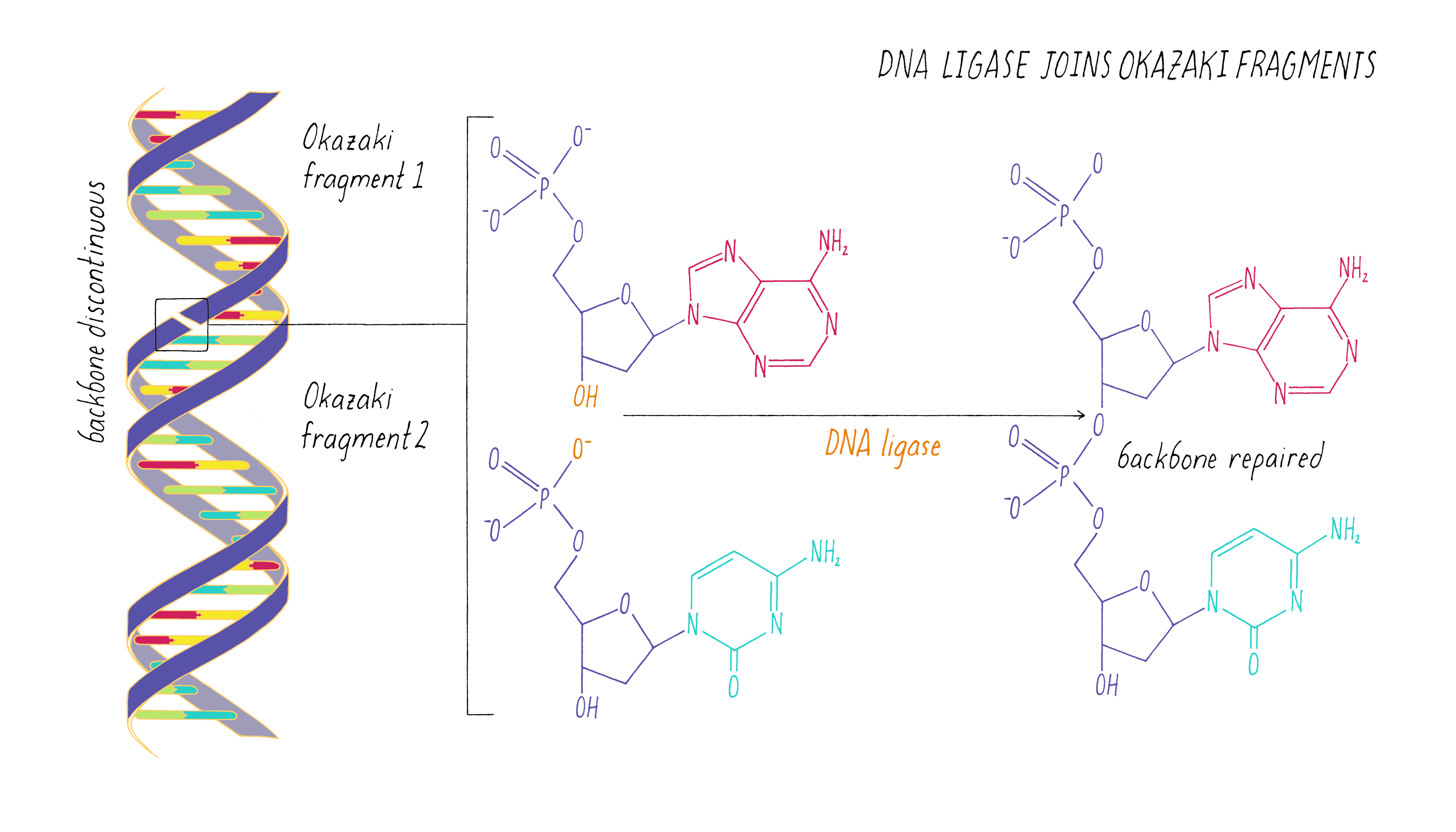
Figures 28–32 show enzymes working in different locations on the leading and lagging strands. However, these, and additional, proteins are located in close proximity at the replication fork. The DNA replication process involves an amazing choreography of these molecular machines as depicted in the animation in Video 7. Even in this video, the speed is slowed down for clarity. The DNA in bacteria can be replicated at a speed of 1000 nucleotides per second. The process operates with extraordinary accuracy in humans (inserting only one wrong nucleotide in 10 billion base pairs). Some degree of imprecision is also important, since it creates the variation necessary for organisms to evolve over time (see Narrative on Mutations by Koshland).
To obtain a more complete explanation of the steps involved in DNA replication, the reader is encouraged to watch the excellent iBiology video by Dr. Stephen Bell (MIT) described in the Resources at the end of this Narrative.
The DNA Code
Francis Crick wrote a letter to his son shortly after deciphering the double helix:
DNA "is a code. If you are given one set of letters, you can write down the others. Now we believe that D.N.A. is a code. That is the order of the bases (the letters) makes one gene different from another gene."
The nature of the code was not known at the time that the DNA double helix was published, but was deciphered in 1961 by Marshall Nirenberg (to be covered in another Narrative). A group of three bases, called a codon, specifies one amino acid. The DNA is copied to a messenger RNA (mRNA) and then the mRNA is read by the ribosome. A triplet code of the four different bases results in 64 possible combinations. One triplet is reserved to instruct the ribosome to "start" the first amino acid of a protein (which is the amino acid methionine). Three triplets are used to instruct the ribosome to "stop" adding amino acids and terminate the polypeptide chain. Since there are 60 remaining codons for the other 19 amino acids, multiple codons can be used to specify the same amino acid. An example of codons that specify a short string of amino acids is shown in Figure 33.
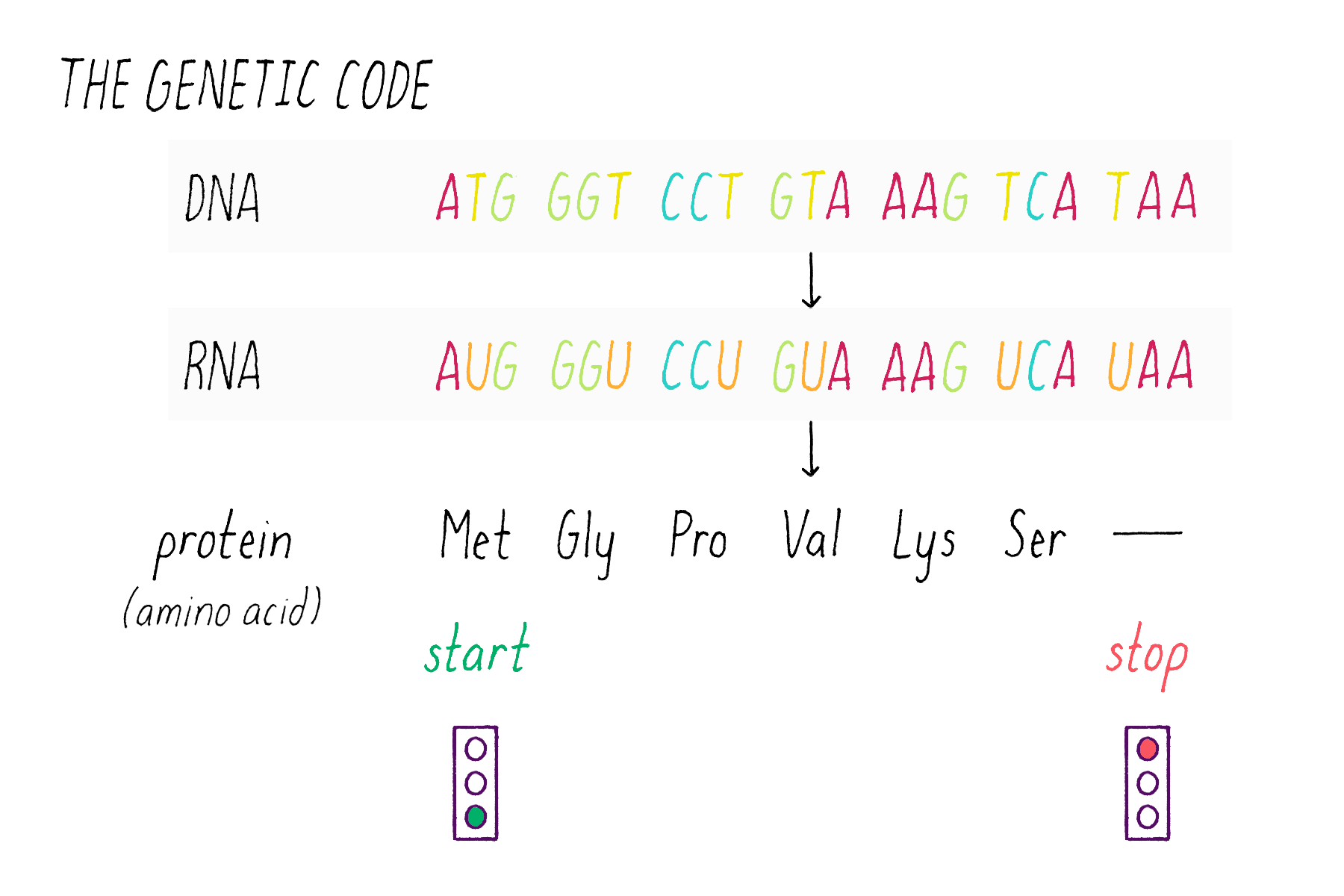
Part III: Frontiers —
DNA nanotechnology
Eukaryotic chromosomes are long, linear DNA molecules; bacterial chromosomes are frequently circles of DNA. However, scientists realized that the simple rules of DNA base pairing could be exploited to create much more complex shapes than are found in nature. Ned Seeman, in 1982, first began exploring the use of DNA as a building material for applications in nanotechnology. Today, DNA nanotechnology has become a booming area of research, attracting both engineers and biologists.
The basic principle of DNA nanotechnology is based upon creating branch points that connect multiple strands of DNA. Biological DNA consists of two strands that are fully complementary, creating a linear molecule. However, four strands of DNA that are partially complementary can interact in the manner shown in Figure 34. Two of the DNA strands act as "staples" that bridge two non-complementary DNA strands that normally would not interact. With this simple design, a simple 2D shape is created: a rectangle of two tightly packed helices. Staples also can connect more than two segments of DNA and thus create more complicated 3D shapes.
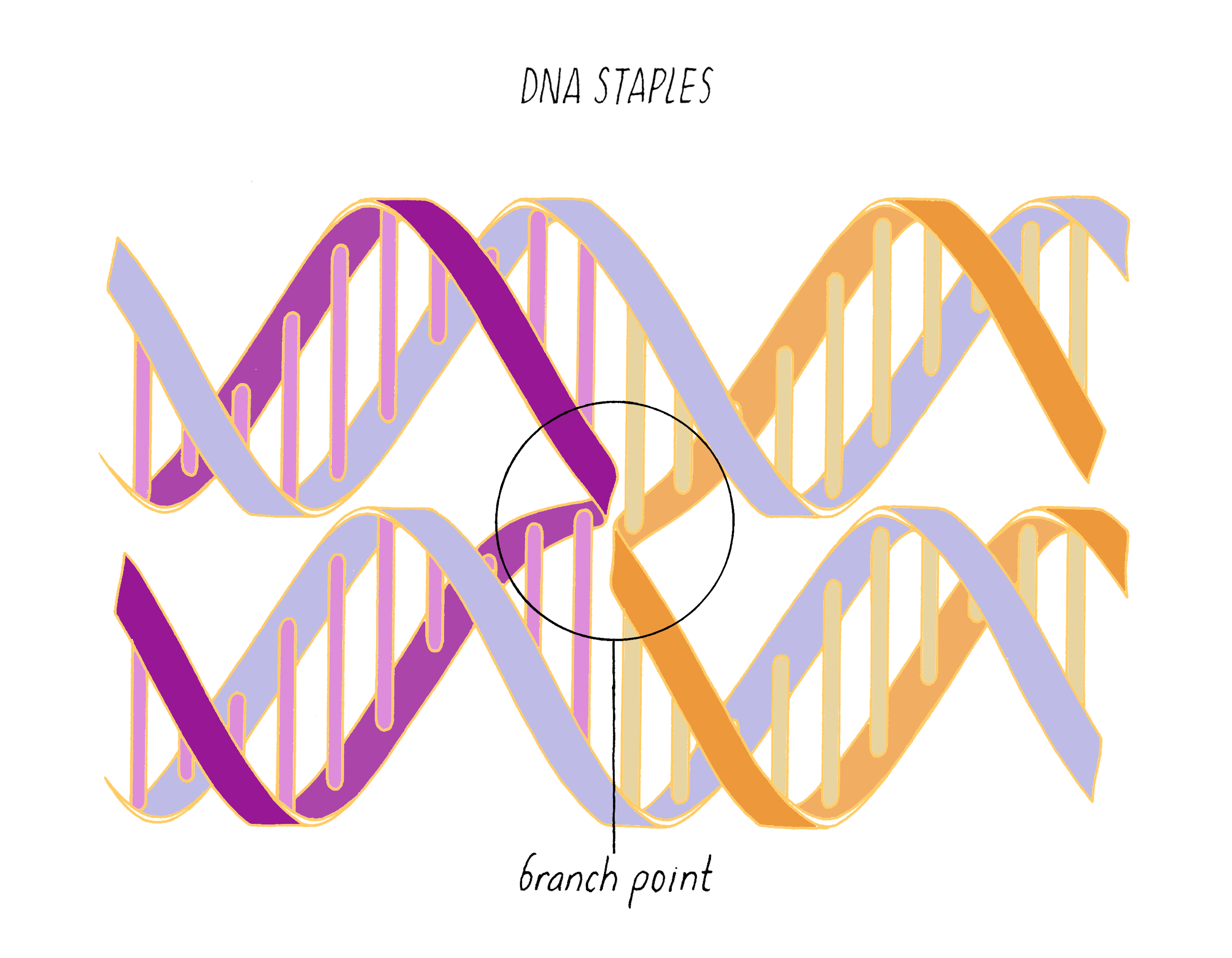
DNA Origami
A popular method of fabricating 3D DNA nanostructures is called DNA origami, which was invented by Paul Rothemund in 2006. To showcase this nanoscale design, Rothmund made smiley faces out of DNA, which were featured on the cover of Nature, the same journal that published the trio of pivotal DNA structure papers from Watson/Crick, Franklin, and Wilkins in 1953 (Figure 35). Keep in mind that these DNA smiley faces are only ∼100 nm in diameter, which is much smaller than a typical human cell (30,000 nm in diameter).

DNA origami involves mixing two components: (1) a very long, single-stranded "scaffold" strand that on its own just forms a very floppy circle, and (2) a mixture of short oligonucleotides of single-stranded DNA (staples) that are complementary to different regions of the scaffold strand. When the staples and scaffold form helices, different parts of the circle are brought together, folding it into a very specific shape (hence the whimsical name "origami"). One advantage of the origami approach is that different shapes can be created from one scaffold with different oligonucleotide staples (Figure 36).
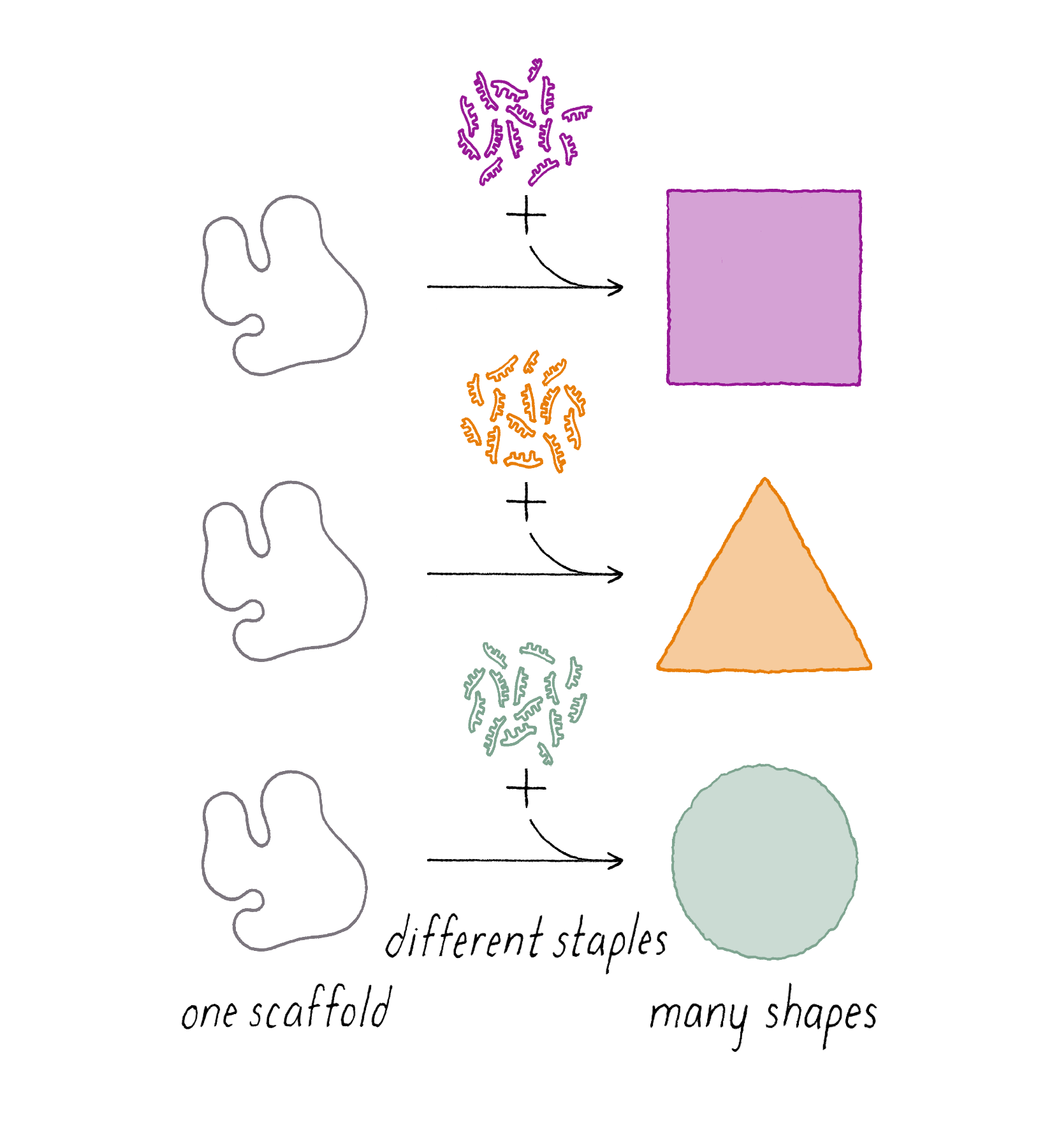
The actual strand-routing diagram of a DNA origami design might look something like that shown in Figure 37:
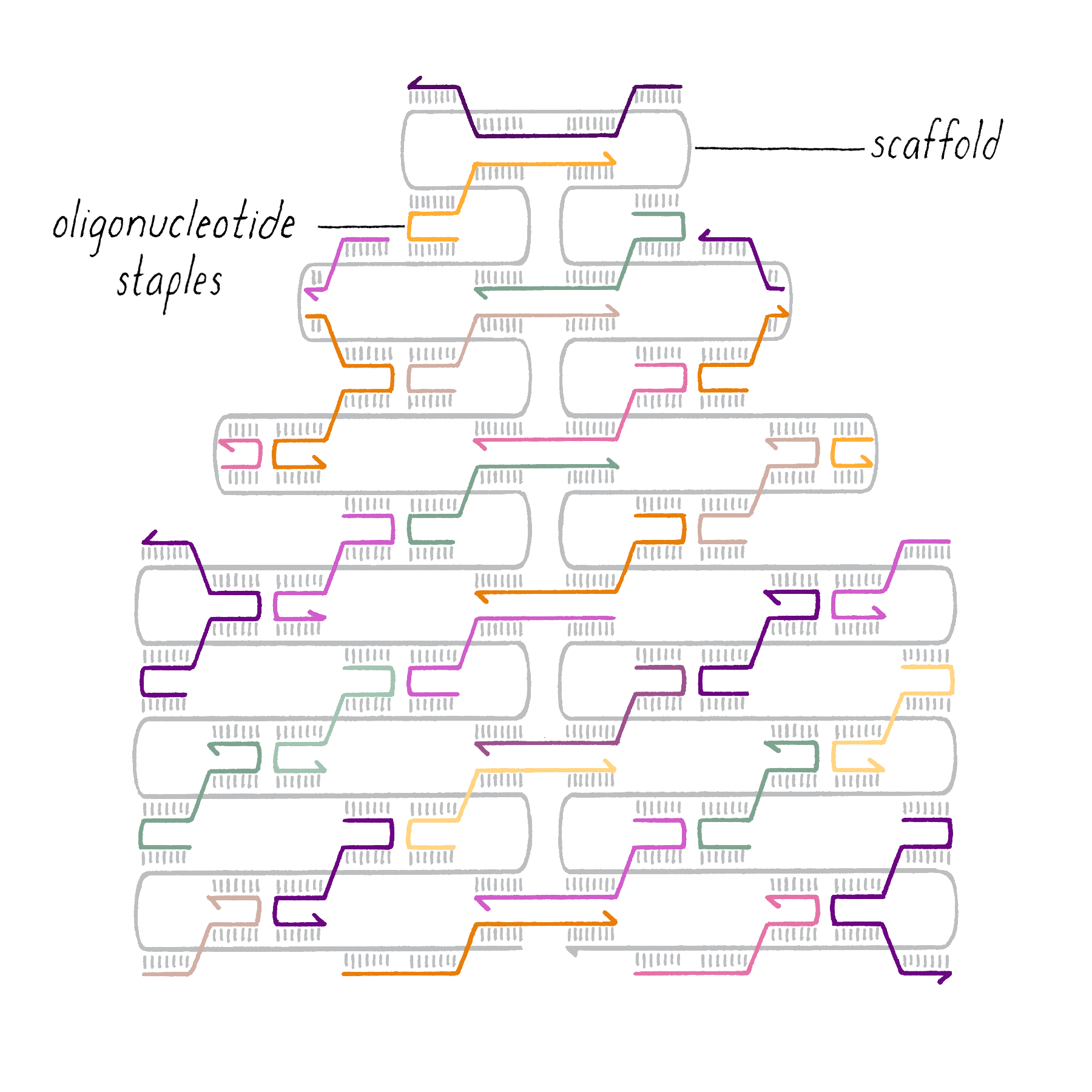
So how did Rothemund make the DNA smiley face? The process involves two important steps.
1. Obtaining DNA
The genome of a small single-stranded DNA virus (∼8000 nucleotides) is typically used as the scaffold. The virus can be prepared in large quantities and its DNA purified. The custom-designed DNA staples are made by humans, and not nature. Many commercial companies use automatic machines that execute a sequence of chemical reactions to synthesize single-stranded DNA with any custom sequence. Today, scientists design a specific DNA sequence on their computer (up to a few thousand nucleotides at a cost of ∼10 cents per base), send in the order electronically to the DNA synthesis company, and a tube of billions of identical single-stranded DNA molecules arrives by courier to their lab bench a few days later (Figure 38). Making custom DNA sequences has certainly become very easy. On the other hand, designing the sequences of ∼200 DNA staples that will produce a specific shape is not trivial. However, computer software (cadnano; https://cadnano.org/) is available to facilitate this process.
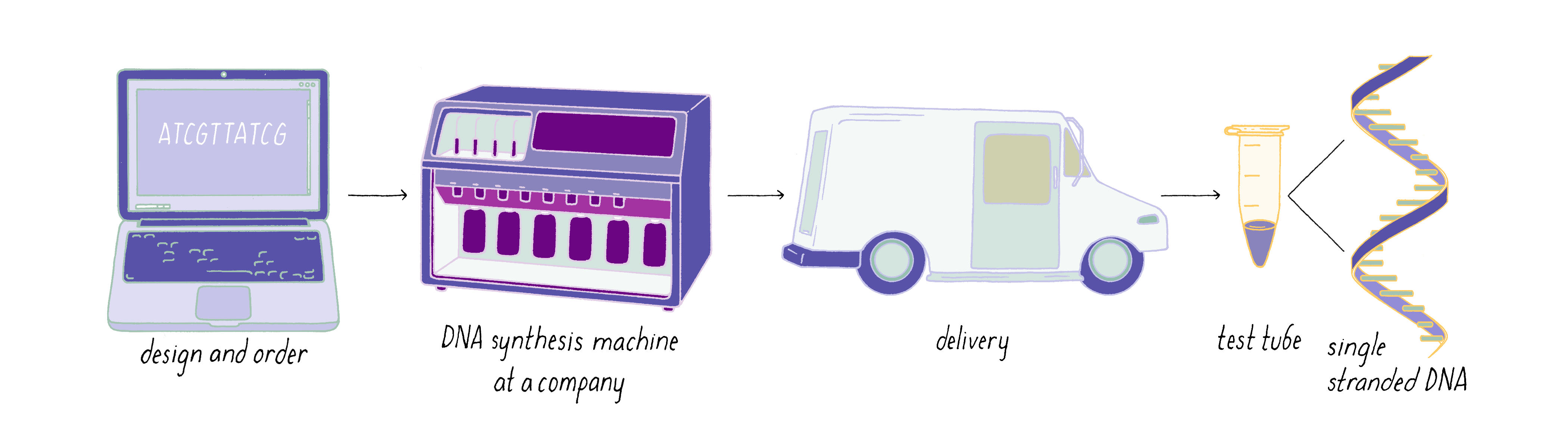
2. DNA Hybridization.
The single-stranded DNA scaffold and staples then need to be combined. The DNA mixture is first heated to break apart any preexisting base pairs, and then cooled down to room temperature to allow base pairing between the scaffold and staples, as depicted in the animation in Video 8.
In the case of Rothemund’s smiley faces, ∼200 distinct DNA oligonucleotide staples were combined with one long (∼7000 nucleotides) single-stranded DNA scaffold. Helices in adjacent rows of the smiley face are connected through branch points created by the DNA staples (Figure 39).
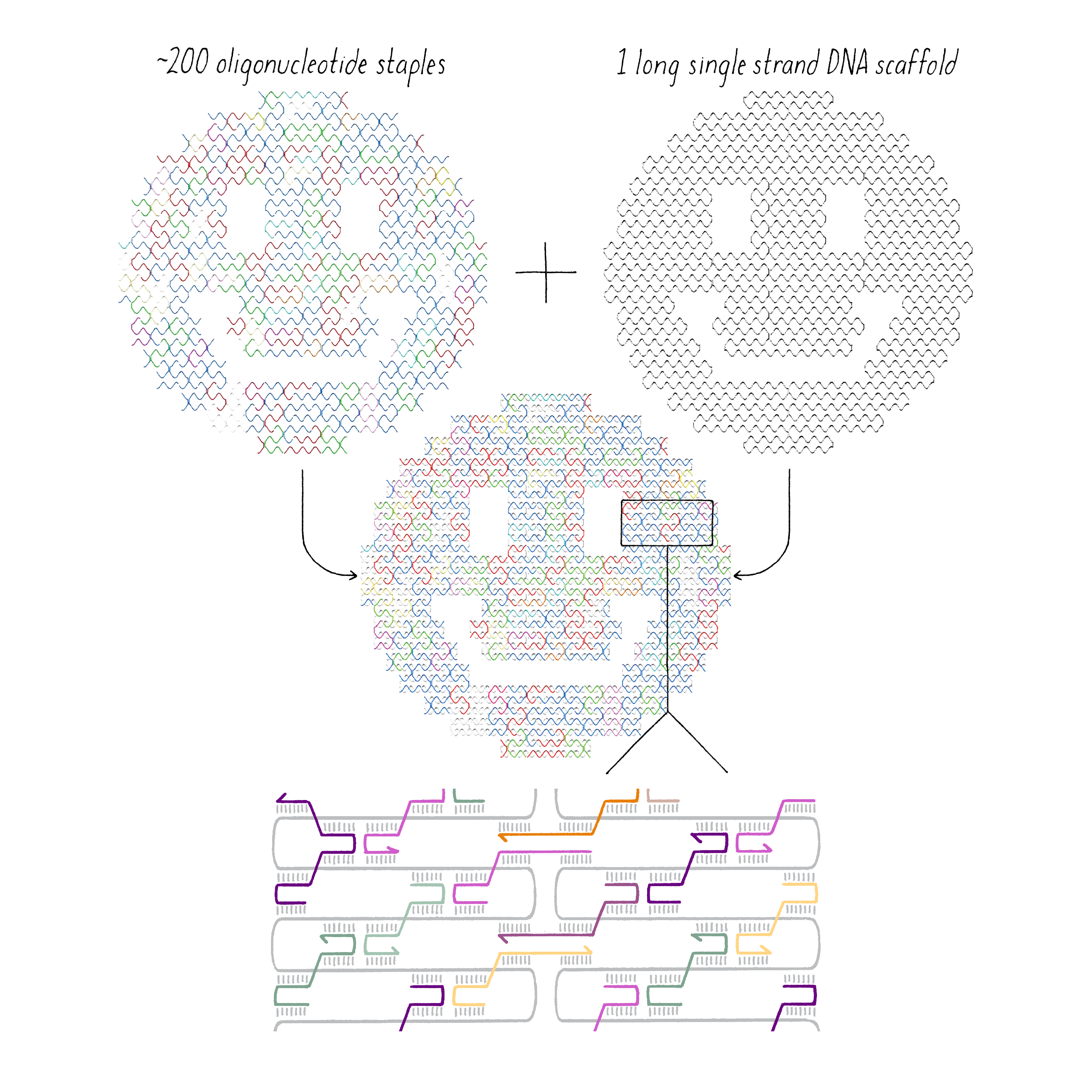
The last step is checking whether the DNA origami process actually worked. As the structures are small (one ten-millionth of a meter), one needs a high-resolution method like electron microscopy or atomic force microscopy to image these structures, as shown in Figure 40.

A DNA Origami Nanorobot that Inhibits Cancer Cell Proliferation
DNA origami is being used in many creative ways. One example is a DNA origami nanorobot was designed to halt the division of cancer cells (see the Guided Paper by Shawn Douglas, Ido Bachelet, and George Church). The nanorobot carries a "payload" (an inhibitory antibody), which it delivers if, and only if, it encounters a cell with a molecular signature characteristic of cancer.
The nanorobot consists of two halves that come together to form a barrel. The two halves are closed with two pairs of complementary DNA strands placed at the mouth of the barrel (Figure 41). The middle of the barrel contains growth-inhibiting antibodies that are attached to the DNA. When the antibodies are trapped inside of the barrel, they cannot interact with cells; as a result, the DNA nanorobot in the closed barrel form is harmless.
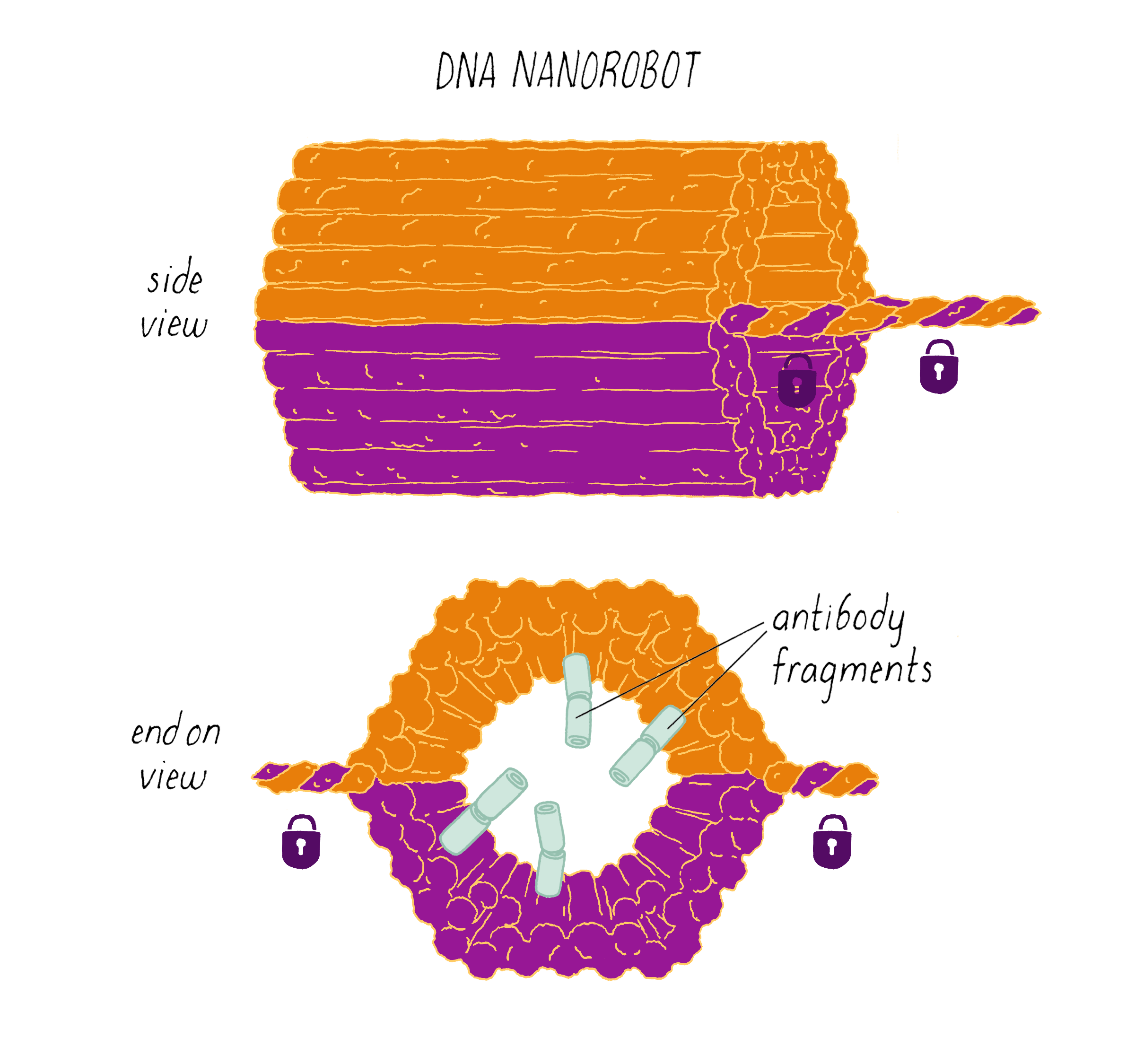
The trick, then, is to get the cancer cell to open up the nanorobot and expose the inhibitory antibodies. To accomplish this, Douglas developed a clever unlocking mechanism. One DNA strand of the pair that keeps the barrel closed has a special capability of binding to a specific protein expressed on a cancer cell. This type of DNA sequence is called an aptamer. When its protein partner is present, the aptamer will let go of its complementary DNA strand and wrap around the protein instead (Figure 42).
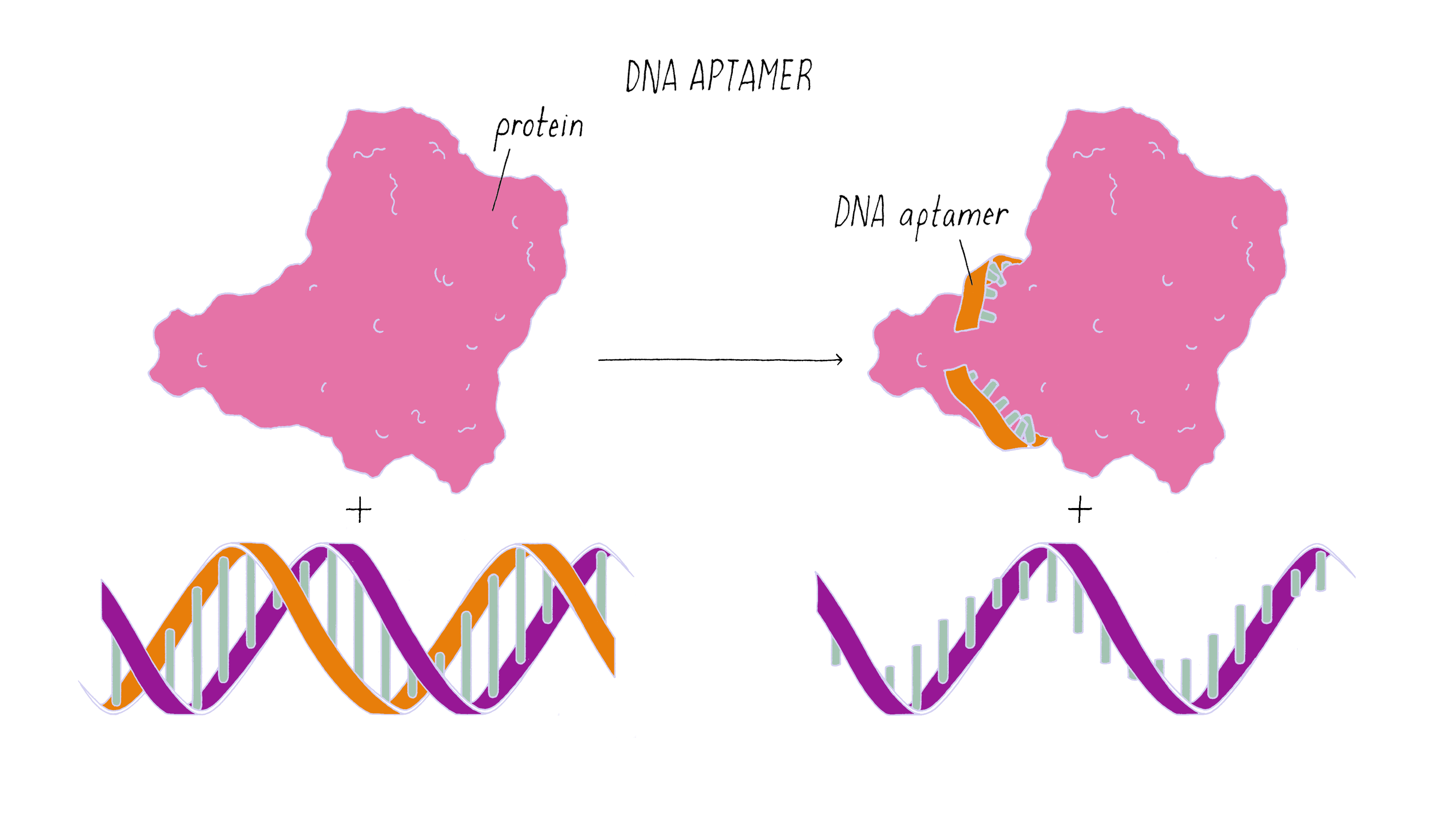
The DNA aptamer provides an "intelligent" lock that keeps the DNA nanorobot closed unless it encounters a protein from a cancer cell (Figure 43). The protein on the cancer cell acts as a "key" that unlocks and opens up the two halves of the nanorobot, exposing the payload inside.
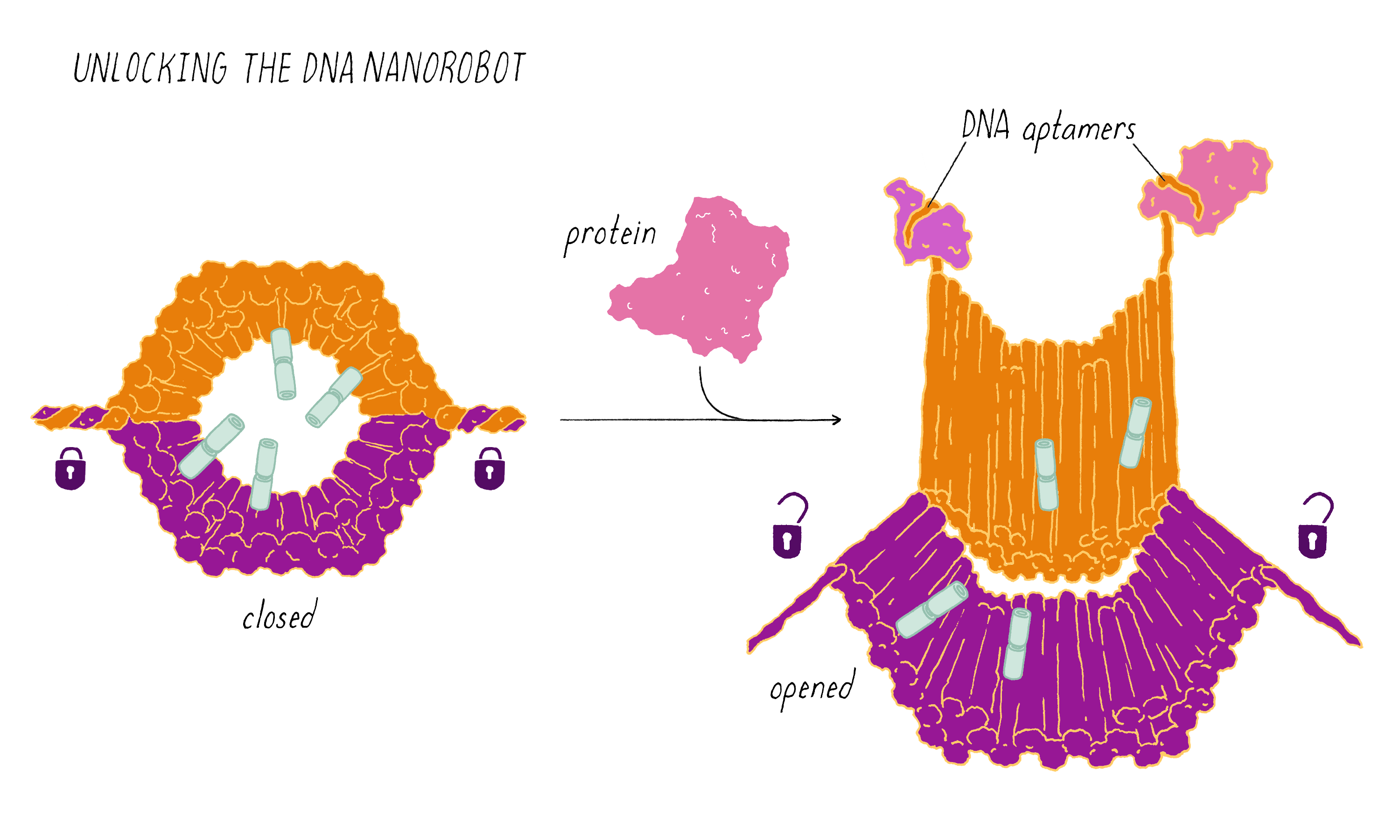
The nanorobot also can be designed to be extra smart, and open up if, and only if, it detects two different proteins instead of one. In this case, two different DNA aptamers, which recognize different proteins, are deployed on either side of the barrel mouth. If one aptamer binds its partner protein but the second aptamer does not, then the barrel will remain closed since only one side is unlocked. Both proteins must be present to unlock and fully open the barrel. This dual recognition aptamer system offers improved specificity for a target. For example, a combination of two cell surface proteins better defines a unique molecular signature of a cancer cell than a single protein.
With these design principles in place, Douglas built a nanorobot that would battle leukemia cells (white blood cell-derived cancer) (Figure 44). The nanorobot’s payload was an antibody that inactivates a cell surface protein that promotes cell proliferation. The opening and closing of the nanorobot was controlled by two DNA aptamers that recognize two different cell surface proteins that are abundant on leukemia cells but not on normal white blood cells. Tests of the nanorobot (see the Guided Paper) confirmed that it opens efficiently in the presence of leukemic cells but not normal white blood cells.
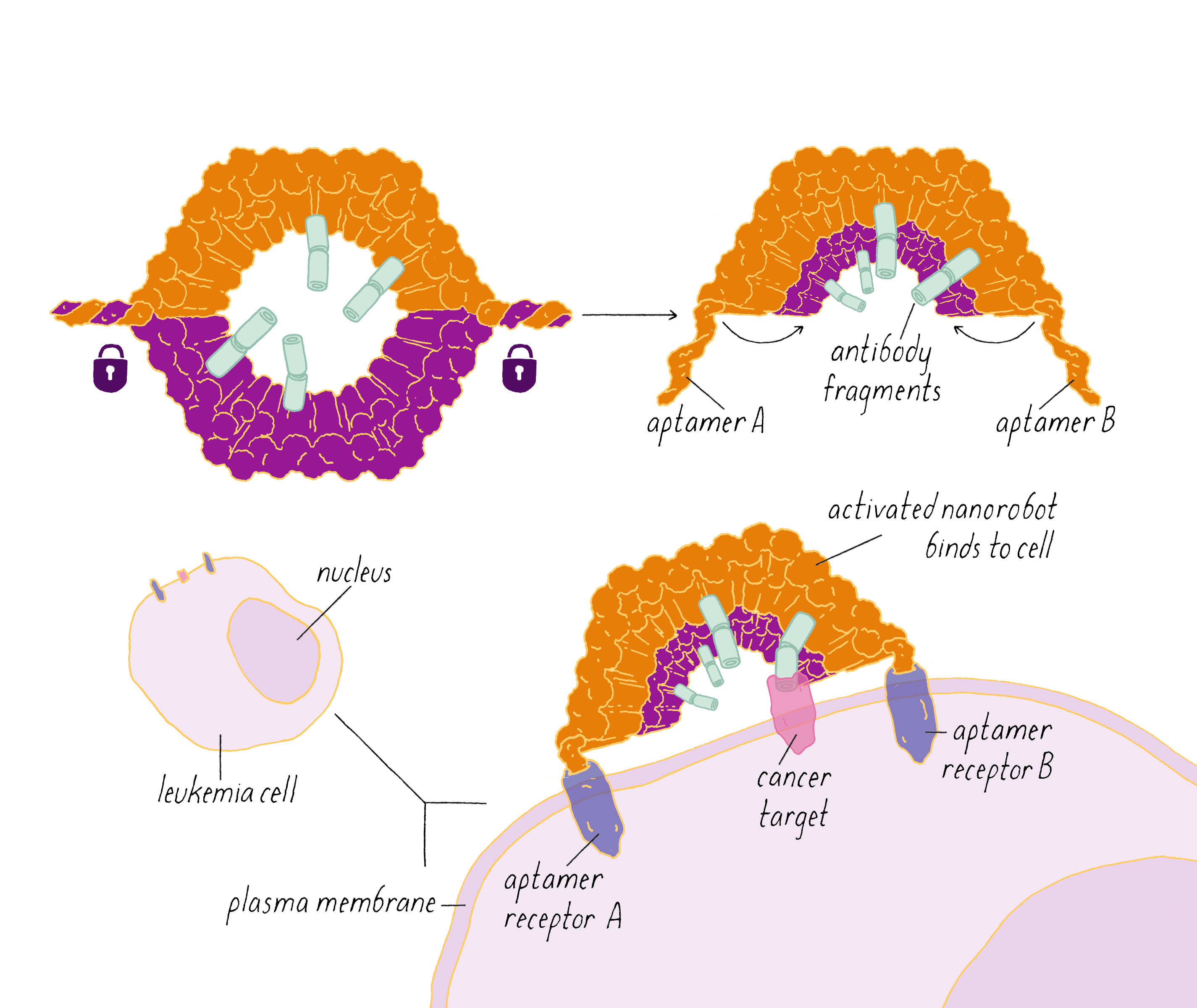
Finally, the nanorobot was ready for testing with leukemia cells growing in cell culture. Three versions of the nanorobot were tested. The "gated" nanorobot had the dual aptamers designed to open up in response to encountering two specific proteins on the surface of the leukemic cells. Two other nanorobots were tested to assess how well the cell-specific locking mechanism worked. One was permanently locked using non-aptamer DNA strands. The second was permanently open as it lacked locking strands. The results are shown in Figure 45. The "always locked" nanorobots had no or little effect on cell division at any DNA nanorobot concentration. In contrast, the "always unlocked" nanorobot inhibited cell division with increasing concentrations of the nanorobot. The "gated" nanorobot also inhibited cell division, indicating that it must have opened up when it encountered the cancer cells. While the DNA nanorobot has not yet been used to treat cancer patients (only cancer cells growing on a dish in the lab), it provides an example of how "smart" nanoscale devices can be created with DNA origami.
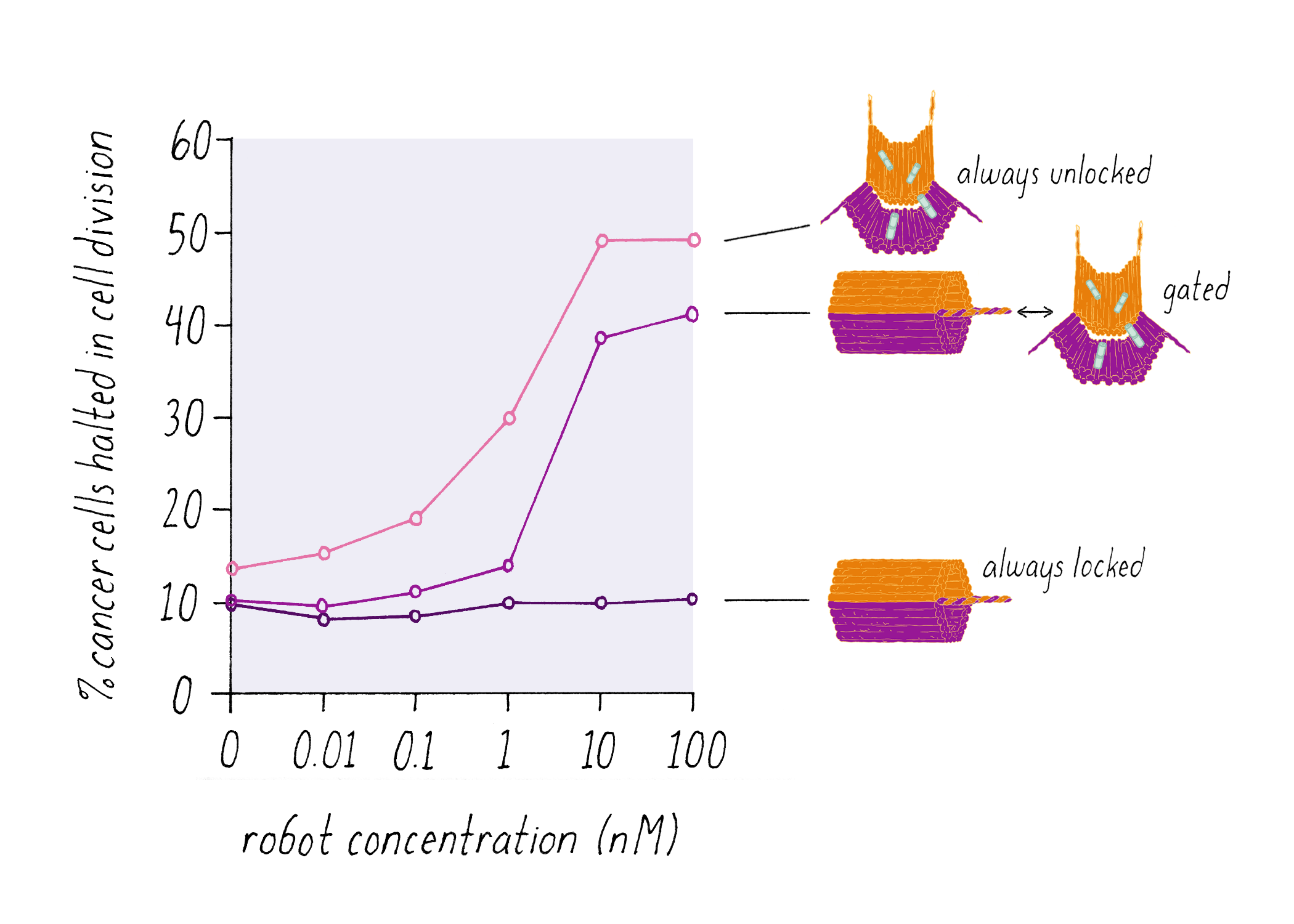
Explorer’s Question: Why do you think that the growth inhibition curve for the "gated" nanorobot is shifted to the right of the "unlocked" nanorobot?
Answer: The rightward shift of the curve indicates that the gated nanorobot is less efficient in inhibiting cell division; for example, at 1 nM concentration, the "gated" nanorobot was ineffective while the "always unlocked" nanorobot had an effect. This result indicates that the gate might be a bit "sticky" and that not all nanorobots open up when they encounter the cancer cells.
Explorer’s Question: If the "gated" nanorobot is less efficient, why not just use the "unlocked" nanorobot for inhibiting cancer cells?
Answer: There is a trade-off between efficiency and specificity. It is desirable to kill the leukemic cells, but undesirable to react with normal white blood cells or bone marrow cells that make white blood cells. The "always unlocked" nanorobots, while more efficient, have the potential to affect normal cells. The "gated" nanorobots provide specificity by targeting the inhibitory payload antibodies to the cancer cells.
Explorer’s Question: Why are only 50% of the cancer cells inhibited in their growth?
Answer: The graph suggests the effect of the nanorobots reaches a plateau at ∼50% growth inhibition (although it would be useful to have additional data points to establish this conclusion in a more convincing manner). There are several possibilities. First, it is possible that the exposure time (3 days) of the nanorobots was too short and that greater inhibition could be achieved with a longer incubation time. Second, the nanorobot inhibition may be partial; 50% of the cancer cells escape nanorobot inhibition and divide. Third, a subset of cancer cells may be resistant to the nanorobot (resistance is a common property of cancer cells; see Narrative on Cancer by Mike Bishop). This third possibility could be confirmed by exposing cancer cells to nanorobots and selecting the survivors. If the survivors are re-exposed to nanorobots and are unaffected, then they are a resistant subpopulation. As an example of resistance, cells might have stopped producing one the protein "keys" needed to unlock the nanorobot.
Explorer’s Question: What other controls could you imagine performing to better understand the specificity of the nanorobots?
Answer: In their paper, the authors showed that the "gated" nanorobot did not bind to normal white blood cells, but it might be useful to test for functional effects as well. For example, T cells (a type of white blood cell) can be induced to divide in response to an invading organism. Does the nanorobot affect the division of T cells? One also might want to know if the "gated" nanorobot opened in response to the correct aptamer-targeting proteins on the leukemia cells. Using current CRISPR gene editing technologies (see Narrative by Barrangou), one can disrupt the genes for these two proteins in these cancer cells. In these gene-edited cells, the "gated nanorobot" should not be able to open and should not inhibit cell growth.
Undergraduates and DNA Nanotechnology
Does DNA origami and nanoscale biomolecular engineering sound cool? But alas only accessible to PhD scientists? Think again. Now undergraduates are getting into the game. Every year, undergraduates from around the world are designing amazing biomolecule-based devices, which include nanoscale robots, computers, and therapeutics, in a competition called BIOMOD. The selected teams of undergraduates then meet in person in San Francisco to share their ideas and potentially win a prize. You can learn about some of the amazing DNA origami ideas that undergraduate students have thought of in Video 9.
Closing Thoughts
B-DNA and A-DNA, the two forms of DNA identified by Rosalind Franklin, could equally stand for the two major epochs of biology: "Before" and "After" the DNA double helix. This turning point raised a new round of fundamental questions about living organisms—How does DNA replicate? What is the nature of the genetic code? How is information transferred from the nucleus (where DNA resides) to the cytoplasm (where proteins are made)? What is the nature of mutations and how is DNA repaired? How do the long DNA strands (∼2 m in total length) get packaged into a small nucleus? We now have general answers to these questions, although important details remain obscure. And new questions have recently arisen that await a next generation of biologists, such as you, to answer. Only 1% of the DNA in the human genome codes for proteins. What is the rest of the DNA for? The genomes of human and chimpanzees are 99% identical in their DNA sequences. How are such different creatures produced through such a small difference in their DNA? Worms and humans both have 19,000–20,000 protein coding genes. What makes us so much more complex? The DNA double helix is like the Mona Lisa of biology—beautiful and mysterious. Much remains unsolved about DNA.
The discovery of the DNA double helix also is perhaps the most told story in biology, having been covered in books, plays, movies, and editorials. It is frequently viewed from the angle of scientists seeking fame and the Nobel Prize, which led to acrimony. While competition is a part of human nature, and scientists indeed are very human, this portrayal fails to capture the science and motivations leading to the discovery of the double helix. Watson and Crick were not thinking of the Nobel Prize. Crick, bright as he was, must have been concerned primarily about finishing his PhD and graduating. Watson, as brash as he was, produced an undistinguished PhD thesis and was likely thinking about proving himself as a scientist. Even after the double helix was published, Crick apparently did not think that the work was prize-worthy until someone casually mentioned the Nobel Prize to him in 1956 (from A Mad Pursuit, see reference list). So what drove them, if not fame? I would argue it was the same factor that drives most scientists: a burning urge to solve a problem. Watson and Crick possessed this urge, they fed each on that passion, and that drove them to success.
In addition to their drive and sense of urgency, a second factor in Watson’s and Crick’s success was their close collaboration. As highlighted in this Narrative, their continuous intellectual sparring and close camaraderie allowed the double helix to come into view in just one magical month. Their complementary skills provided the ideas that were necessary for the puzzle to be solved. As Crick said, "if Watson had been killed by a tennis ball, I am reasonably sure I would have not solved the structure alone…" (What Mad Pursuit, see references). Meselson and Stahl (see their Key Experiment on How DNA Replicates) also cite their collaboration as critical for their success. There is a general lesson here for young people reading this Narrative. Find a buddy (or two) with whom you can share ideas, whether it is science, music, business, or something else. Two minds think better than one; your partner can prevent you from getting stuck in an intellectual or creative rut that you might not be able to spot yourself. Success also is more joyful and inevitable difficulties seem less disappointing when working with a friend.
A third element for scientific success, again revealed by the double helix, is freedom of independent thinking. Watson and Crick were junior scientists; they were helped, but not told how to think by their seniors. (Meselson and Stahl also expressed similar sentiments in their Key Experiment.) Scientists, of any age, desire the challenge of independently solving a problem. If successful, they deserve to be recognized for their contribution.
In contrast to the independence and credit bestowed upon Watson and Crick, Florence Bell’s pioneering work on DNA structure is usually credited to William Astbury, her PhD advisor. Bell published six papers with Astbury in Nature, but she was always placed behind Astbury in authorship. Yet, from reading her thesis (which usually provides direct insight into the student), she appears to have been an intellectual driving force in Astbury’s laboratory at that time. Would Bell have continued a career in science if she had been better recognized for her work? Would the University of Leeds have made a key breakthrough on DNA structure before King’s College if she had?
Rosalind Franklin’s desire for independence in her scientific work at King’s College created conflict. Because she was isolated, either by choice or necessity, Franklin was handicapped by the lack of a close collaborator who served as an equal intellectual sparring partner. She operated as a single-strand, lacking the complementary strand that Watson and Crick had in their collaboration. Would King’s College have reached a model of a DNA double helix if Rosalind Franklin’s independence had been embraced and if she and Wilkins collaborated?
The story of DNA reminds us how biases can interfere with scientific progress. Being a female scientist in the era of Bell and Franklin was difficult. Even now, women are underrepresented as academic professors and other senior scientist positions, and there are contemporary stories reminiscent of Bell’s and Franklin’s. The good news is that many scientists are striving to do better and are actively working to help young scientists of all genders and ethnicities to realize their professional dreams.
Finally, I will close by saying a word about stay true to your dreams. DNA structure was neither Watson’s nor Crick’s main project. Crick was working on protein structure and Watson was working on bacterial genetics, among other things. They were blacklisted from working on DNA for a while. They experienced failures along the way. It was not clear whether they would come up with a solution to the DNA structure. If they did, it was not clear that the structure was going to be interesting. Yet, amidst such doubts and uncertainties, they returned to DNA time and time again. DNA inspired them; it filled their dreams and private conversations. They pushed forward based upon the intuition that they were working on something important and the audacity that they, two young scientists, could solve the problem before anyone else. The story of the double helix reveals that dreaming, audacity, intuition, collaboration and perseverance, a set of skills that are not typically taught in the classroom, are ingredients for doing great things.
Acknowledgment
I thank Shawn Douglas, Kara McKinley, and Cynthia Wolberger for their constructive comments.
Dig Deeper
Dig Deeper 1: The chromosome theory of inheritance
Chromosomes came to forefront in the late 19th/early 20th century as cell biologists began to study their structures and behaviors by microscopy. The term chromosome was coined in reference to the subcellular "bodies" (soma in ancient Greek means "body") that were brightly stained with certain chemical dyes (chroma in ancient Greek means "color"). The theory of chromosome inheritance emerged after the rediscovery of Mendel’s long-forgotten 1865 paper on inheritance in 1900 (see Narrative on Inheritance by Tilghman). Theodor Boveri, studying roundworms, noted that chromosome numbers were reduced in half (haploid) during the divisions of the germ cells to produce gametes. Chromosome copy number then was restored to the normal level (diploid) during fertilization, which was consistent with chromosomes carrying copies of heritable material from the mother and father. Sutton, studying grasshoppers, described the pairing of maternal and paternal chromosomes and their subsequent separation during the cell divisions that produce gametes, a process that we now refer to as meiosis (see Video DD1). Sutton clearly articulated the theory by writing, "I may finally call attention to the probability that the association of paternal and maternal chromosomes in pairs and their subsequent separation during the reducing division as indicated above may constitute the physical basis of the Mendelian law of heredity."
Dig Deeper 2: The Bell and Astbury hypothesis that DNA creates a scaffold for proteins, which jointly create a structure for heredity
Bell and Astbury were struck by the close match between the spacing between the bases in DNA (3.34 Å) and the spacing between amino acids in a stretched out polypeptide (3.4 Å). They thought that this very similar spacing must reflect some important connection between the two. Bell said in her thesis, "The most striking attribute of the nucleic acid column is the periodicity of the nucleotides, 3.34 Å, which is equal to the side-chain period of a fully extended polypeptide chain. It is difficult to believe that the agreement is no more than coincidence; rather it is stimulating thought that probably the interplay of proteins and nucleic acids in the chromosome is largely based on this very circumstance." Bell and Astbury went on to speculate that DNA in chromosomes serves as a scaffold to align amino acids from proteins, and that the aligned amino acids might provide the information for heredity. This hypothesis is incorrect, but one must remember that their ideas came at a time when there were many misconceptions about DNA, such as the tetranucleotide hypothesis and the widely accepted belief that genes were proteins.
However, Bell’s general idea that proteins and nucleic acids together form (in her words) "the long scroll of life on which is written the pattern of life" ended up proving prophetic. We now know that many proteins interact with DNA (e.g., transcription factors) and that these interactions allow the DNA code to be read out at the right time and at the right amount. In reality, proteins and DNA (not just DNA alone) determine "the pattern of life."
Dig Deeper 3: Franklin’s Photograph 51
Here, we briefly describe how the pattern of spots in Franklin’s X-ray photograph can be interpreted to provide information on the structure of DNA. Detailed information on the theory of X-ray diffraction is beyond the scope of this Narrative; however, more information can be obtained in the Resource section.
The spots in the photograph are made by X-rays that interact with atoms in the DNA and are scattered, meaning that they diverge from the straight trajectory that the X-ray beam took when it entered the DNA fiber. A spot emerges if many X-rays arrive at this particular location and do so in "phase," which means that the peaks and troughs of the X-ray waves are aligned (constructive interference). If they are misaligned, a spot will not be produced. If atoms are ordered at regular intervals, then many of the scattered X-rays waves hit the photographic film in phase with one another and expose the photographic emulsion. Thus, the spots (or lines) in the photograph provide information on regular, repeating patterns of atoms in the sample. The information, however, is in a curious "reciprocal space," which means that a smaller regular spacing of atoms in the sample produces spots farther from the center.
In photograph 51, one can see 10 equally spaced "lines" from the axis, which are called "layer lines" (Figure DD3). The closest layer line to the center reflects the largest regular repeating pattern of atoms in the DNA. This represents the distance of the regular helical repeat, which can be calculated from its location in the photograph to be 34 Å. The farthest layer line from the center (the arc at the top) reflects the smallest regular spacing in DNA. This is the spacing between bases, which is 3.4 Å (also noticed by Florence Bell in Clue 2 in the Journey to Discovery ). The 10 regularly spaced layer lines are produced by the 10 bases that make up one repeat of the helix. The layer lines also form a distinctive X-shape, signifying a helix. This is due to the zig-zag pattern of the phosphate backbone, as it travels to the left and then to the right across the axis of the double helix. The angle of the X in the photograph reveals the pitch angle of the helix (Figure DD3). The absence of a fourth layer line is a consequence of the slight offset of the two phosphate backbones (also providing evidence of a double helix).
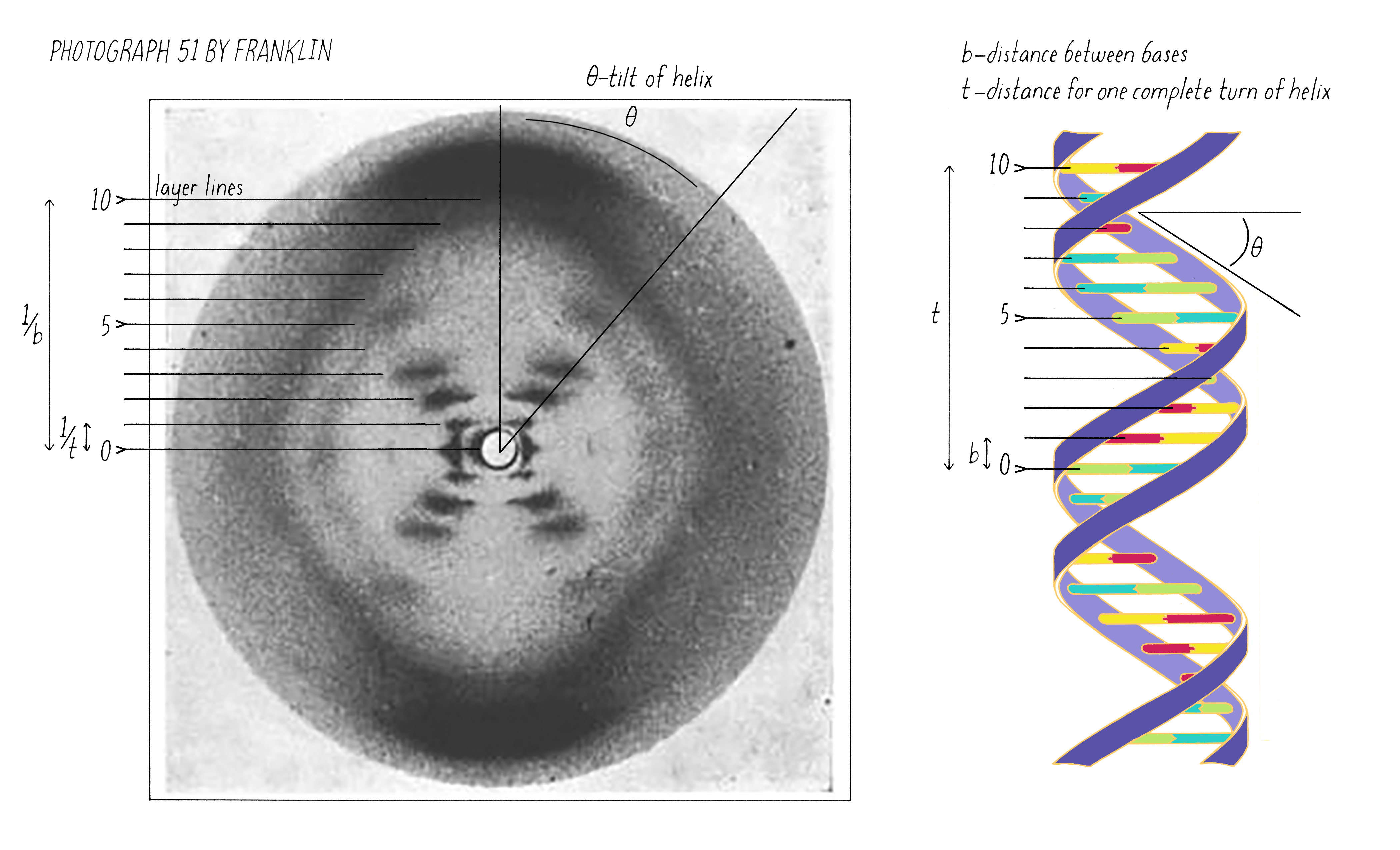
Dig Deeper 4: Different forms of DNA
DNA can adopt different double-stranded structures depending upon the conditions, such as the salt concentration and water content. The B-form is found in living cells and is the form primarily discussed in this Narrative. The A-form of DNA is observed under dehydrating conditions (less water); compared to B-DNA, it is wider, the repeat distance of the helix is shorter, and the bases are slightly tilted. Another helical form is Z-DNA, which forms in a test tube under high salt and with certain base sequences (alternating C-G and G-C pairs). Z-DNA is a left-handed helix (unlike the B- and A-forms) and has a narrower diameter and longer helix repeat than the B-form. Z-DNA may form under some limited circumstances in biological systems.
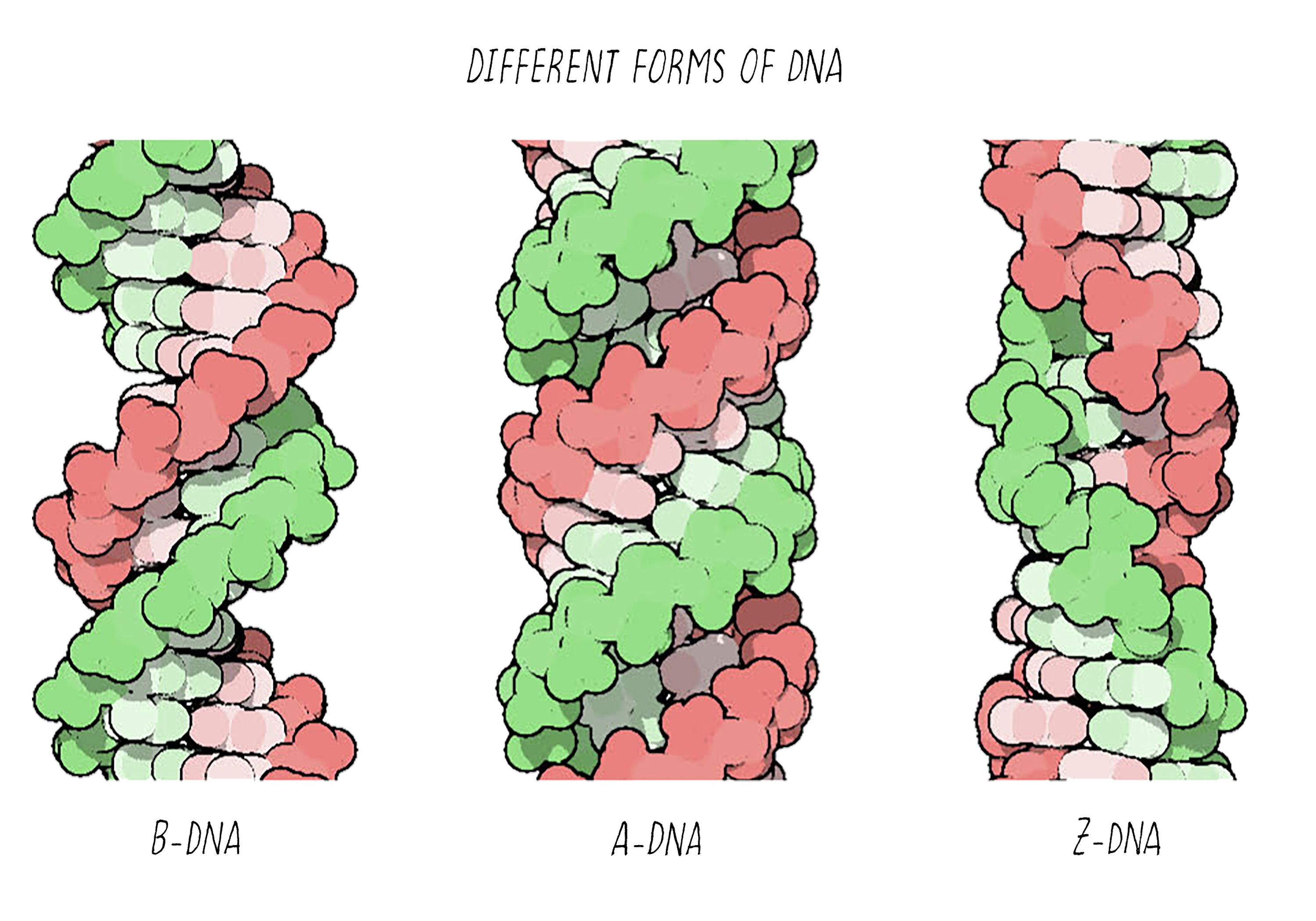
References and Resources
Guided Papers
Watson, J.D. and Crick, F.H.C. A structure for deoxyribose nucleic acid. Nature 1953;171:737–738.
This classic paper showed that DNA has a double helix structure, importantly revealing an elegant and simple mechanism for replication and the inheritance of genes. This research led to Watson and Crick receiving their Nobel prizes in 1962.
DownloadWatson, J.D. and Crick, F.H.C. Genetic implications of the structure of deoxyribonucleic acid. Nature 1953;171:964–967.
This classic paper proposes that the precise sequence of DNA nucleotides is the code which carries genetic information, and that each strand of the DNA double helix can serve as a template to create a new copy, thus allowing the genetic information to passed along from one cell or organism to its progeny. With the previous paper, this research led to Watson and Crick receiving their Nobel prizes in 1962.
DownloadDouglas, S.W., Bachelet, I., and Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012;335:831–834.
This paper describes the design and use of DNA nanorobots to transport molecules to cells, by selectively interacting with cells to deliver signaling molecules to cell surfaces.
DownloadReferences
- Bell, F.O. X-ray and related studies of proteins and nucleic acids. PhD Thesis, University of Leeds, 1939. Available at: https://explore.library.leeds.ac.uk/special-collections-explore/650413
- Chargaff, E., Zamenhoff, S., and Green, C. Composition of human desoxypentose nucleic acid. Nature 1950;165:756–757.
- Crick, F. What Mad Pursuit: A Personal View of Scientific Discovery. Basic Books, 1988.
- Franklin, R.E. and Gosling, R.G. Molecular configuration of sodium thymonucleate. Nature 1953;171:740–741.
- Judson, H.F. The Eighth Day of Creation. Cold Spring Harbor, NY: Cold Spring Harbor Press, 1996.
- Pauling, P. and Corey, R.B. A proposed structure for nucleic acids. Proceedings of the National Academy of Science U.S.A. 1953;39:84–97.
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006;440:297–302.
- Watson, J. D. The Double Helix: A Personal Account of the Discovery of the Structure of DNA. Signet Books, 1968.
- Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, and Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature 1980;287:755–758.
Resources
- iBiology video on DNA replication by Stephen Bell (MIT) entitled "Mechanisms of chromosomal DNA replication: The DNA replication fork." Available at: https://www.ibiology.org/genetics-and-gene-regulation/mechanisms-chromosomal-dna-replication/#part-1
- DNA: Molecule of the month. From PDB-101, the education portal of the protein data bank (PDB). Available at: http://pdb101.rcsb.org/motm/23
This site allows to one to explore different 3D views of DNA in an interactive manner. - The DNA Learning Center. Available at: https://dnalc.cshl.edu/
- Lucas, A. A-DNA and B-DNA: Comparing their historical X-ray fiber diffraction images. Journal of Chemical Education 2008;85:737–743.
- Rosalind Franklin’s X-ray photo of DNA as an undergraduate optical diffraction experiment. Thompson, J., Braun, G., Tierney, L., Wessels, L., Schmitzer, H., Rossa, B., Wagner, H.P., and Dutz, W. Rosalind Franklin’s X-ray photo of DNA as an undergraduate optical diffraction experiment. American Journal of Physics 2018;86:95. https://doi.org/10.1119/1.5020051 Re-creates aspects of helical diffraction with a laser and the spring of a ballpoint pen.
- The Double Helix Discovery. Video by HHMI Biointeractive. Available at: https://www.biointeractive.org/classroom-resources/double-helix
- And associated resources for educators. Available at: https://www.biointeractive.org/classroom-resources/double-helix