Plant Genetics and the Future of Food
Pamela Ronald
 Pam Ronald
Pam Ronald
Pam Ronald, Ph.D., is a professor at the University of California Davis in the Genome Center and the Department of Plant Pathology, a founding faculty director of the Institute for Food and Agricultural Literacy and a GCHERA World Agriculture Prize Laureate. In 2012, Ronald, Dave Mackill, and Kenong Xu received the 2012 Tech Award 2012 for innovative use of technology (described in the Journey to Discovery) to benefit humanity. Ronald has engaged the public about plant breeding through the book Tomorrow's Table: Organic Farming, Genetics and the Future of Food co-authored with her husband, Raoul Adamchak
Summary
By the year 2100, the number of people on Earth is expected to increase by ~50%, placing increasing demands on food production in a time when a changing climate is predicted to compromise crop yields. Feeding this future world requires scientifically informed innovations in agriculture. Throughout human history, genetic improvement of plants has led to the development of food crops that are useful to farmers and that are nutritious/pleasing to consumers. Here, I will tell the story of the discovery of a rice gene that confers tolerance to flooding and describe how introduction of that gene into rice, a staple food for half the world’s people, has helped farmers in South and Southeast Asia mitigate rice crop failure during prolonged flooding. I will also highlight efforts to modify rice to produce pro-vitamin A with the goal of diminishing the devastating effects of vitamin A deficiency, which is common in many parts of the world. This Narrative provides useful case studies of how international teams of scientists can use knowledge of basic plant biology to address major agronomic challenges faced by farmers and society. I will also explain the similarities and differences between conventional versus modern methods of genetic improvement of plants to inform readers about several of the latest genetic tools available for crop improvement.
Learning Overview
Big Concepts
Natural selection gave way to human selection of plant traits approximately 10,000 years ago with the inception of agriculture. Today, more precise genetic tools are available for crop development, and it is imperative that today’s students, and society as a whole, understand the continuum of plant genetic improvements and how they have been applied in crop production.
Bio-Dictionary Terms Used
5′ and 3′ of DNA, allele, amino acid, amylose, chromosome, DNA, DNA clone, DNA/RNA hybridization, endosperm, food security, genome editing, gene, gene expression, homozygous/heterozygous, locus, mitosis, meiosis, mRNA, mutagenesis, nucleotide, ovule, pathogen, photosynthesis, phospholipid, promoter, restriction enzyme, start and stop codons, stamen, transcription factor, and vitamin
Terms and Concepts Explained
Agrobacterium, bioengineered food, crossing-over, certified organic food, DNA marker, genetic map, GMO, golden rice, grafting, marker-assisted breeding, recombination, northern blot, pollination, sustainable agriculture
Introduction
-
Kenong Xu grew up in a village in China that suffered from rice crop failure due to flooding. Later in life, he would team up with Pam Ronald and David Mackill to identify a gene that aids rice to survive under conditions of prolonged submergence.
Part I: Journey to Discovery - The Search for a Rice Gene that Controls Tolerance to Submergence
-
Prolonged flooding can kill rice and can cause large-scale crop loss in many parts of Asia. In India and Bangladesh alone, four million tons of rice, enough to feed 30 million people, are lost to floods each year.
-
A scientific team sought to identify a gene that makes rice more tolerant to submergence under water and then to introduce that gene into commercial rice varieties.
-
Gene mapping strategies were used to identify the submergence tolerance gene. A genetic map provides an approximate location of the gene on a chromosome and a physical map allows one to pinpoint the gene and its DNA sequence.
-
Genetic mapping was first introduced by Thomas Hunt Morgan and his student Alfred Sturtevant. This method relies on the phenomenon that homologous chromosome can exchange large segments of a DNA, a phenomenon known as “recombination” or “crossing over.”
-
To understand recombination, the basics of meiosis and the terms chromosome, homologous chromosomes, chromatid, gene, and allele are reviewed.
-
The greater the distance between two genes on a chromosome, the more likely that a recombination event will occur between them during meiosis. Thus, by measuring the frequency of recombination, one can obtain an approximate distance between genes and thus create a genetic map. Distances between genes in this map are measured in “centimorgans,” in honor of the scientist who uncovered this principle.
-
Scientists use “DNA markers” as landmarks on a chromosome. These are regions where the DNA varies between individuals of a species and can be used to distinguish between two parents or between parents and offspring. Through genetic mapping, the unknown submergence tolerance gene was mapped between two DNA markers.
-
DNA cloning was used to create a “physical map” between the two markers that flanked either side of the submergence tolerance gene. The physical map is the precise DNA nucleotide sequence along the chromosome.
-
By analyzing the DNA sequence in the physical map, several candidate genes were found. Only one gene (called Sub1), however, was turned on when the rice plants experienced flooding. This gene was also found in submergence tolerance rice varieties but not in rice that were submergence sensitive.
-
To prove that Sub1 was indeed responsible for submergence tolerance, the gene was introduced into submergence intolerant plants. After bioengineering, the rice became submergence tolerant, demonstrating that the Sub1 gene was sufficient to confer this property.
-
Sub1 is a transcription factor that suppresses rice shoot elongation during flooding. This waiting mechanism allows the rice plant to conserve energy until the flood subsides and conditions are good for reinitiating growth.
-
After its discovery, the Sub1 gene was introduced in rice varieties that are commonly used by farmers throughout Southeast Asia through marker-assisted breeding. In 2017, 6 million farmers grew Sub1 rice, helping to minimize their loss of rice crops due to flooding.
Part II: Knowledge Overview - Old and New Strategies of Plant Genetics
-
Farming began in ~10,000 BC. These farmers were early plant breeders who selected for traits in wild plants to generate domesticated plants that were suitable for agriculture.
-
Two examples of ancient artificial selection are presented – the development of modern corn from the wild grass called Teosinte in Mexico and the cultivation of “sticky rice” in Southeast Asia.
-
Plants produce progeny seed through self- or cross-pollination. Plant breeders have learned to control the pollination process. Controlled pollination was used by Gregor Mendel in his famous experiments on heredity in peas and in modern times is used to make hybrids between two varieties of plants.
-
Marker-assisted breeding is a modern method in which progeny with desired traits from a cross-pollination between two plants can be identified and selected. Marker-assisted breeding uses DNA analysis to identify a desired or undesired trait, which is more precise and faster than conventional approaches.
-
Induced mutagenesis is used to increase the frequency at which new traits develop. With induced mutagenesis combined with artificial selection, the rate of plant evolution for crop development is accelerated.
-
Grafting is a process that brings two different species physically together, for example, the roots (rootstock) of one type of walnut tree with the upper part (scion) of another.
-
Bioengineering uses a soil bacterium called Agrobacterium tumefaciens to transfer a plasmid containing a gene of interest into a plant cell and integrate it into the plant genome. Bioengineering additionally allows the transfer of genes from organisms other than plants.
-
Bioengineering introduces a specific gene(s) while conventional breeding mixes large numbers of genes, producing complex phenotypes.
-
Genome editing using CRISPR-Cas allows scientists to specifically change a single gene or insert a gene at a targeted chromosomal location.
-
Nearly all of the food that we eat today have been genetically altered by humans in some way. The use of the terms genetically modified organism, bioengineered, and certified organic are discussed.
Part III: Frontiers - Golden Rice, a New Crop to Fight Vitamin A Deficiency
-
Vitamin A is a chemical needed for human growth, development, vision, and immune function. The World Health Organization estimates that ~250 million preschool children are vitamin A deficient and that 250,000 to 500,000 vitamin A-deficient children become blind every year, half of them dying within 12 months of losing their sight. VAD is most prevalent among the poor in Asia and Africa.
-
Humans acquire β-carotene (also called pro-vitamin A) from diet and the liver converts β-carotene into vitamin A. β-carotene is absent in rice grain, which constitutes the prevalent diet for much of the world.
-
β-carotene is produced by a biosynthetic pathway, which is a chemical production line of enzymes that produces specific chemicals that are essential for life.
-
Golden Rice is a genetically engineered rice that produces β-carotene in the rice seed. This was accomplished by introducing two genes, one from corn and one from bacteria, which encode enzymes that enable rice seeds to produce β-carotene. The “golden color” of the rice arises from β-carotene, which also gives carrots their orange color.
-
Golden Rice, a low-tech, sustainable, publicly funded effort, holds promise for minimizing VAD around the world and can complement other approaches, such as the promotion of home gardens with vitamin A-rich crops.
Closing Thoughts
-
With 11.2 billion people on Earth by 2100 and increasing environmental pressures such as climate change, sustainable agriculture will become increasingly important. Genetic improvement of crops will play a central role in sustainable agriculture, along with social, economic, and environmental policies.
Guided Paper
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., Ismail, A.M, Bailey-Serres, J., Ronald, P.C., and D. J. Mackill. (2007). Sub1A is an ethylene-responsive-factor-like gene that confers submergence tolerance to rice. Nature, 442: 705–708.
This paper describes the discovery of a gene that controls the ability of plants to resist being submerged. As a result, new strains of rice can be produced that are protected against otherwise damaging floods, increasing food security for farmers.
DownloadThis is the paper featured in the Journey to Discovery.
Activity
Learn the principles of agarose gel electrophoresis yourself and run your own simulated gels in the Activity of Gel Electrophoresis by Douglas.
Introduction
Kenong Xu, the graduate student who made the key breakthrough in this Narrative’s Journey to Discovery, lived with his parents, brother, and two sisters in a small house in Yang-Jia-Shan, a small village in China. The family raised farm animals (hens, ducks, geese, and pigs) to add variety to their main diet of rice, which was served for breakfast, lunch, and dinner.
The 30 families in the village shared responsibility for growing rice. Early in the spring, before school, Kenong would help the women transplant the seedlings into the wet paddy, bending close to the ground to be sure that each seedling was unbroken and its roots buried in the mud (Video 1). Throughout the summer, he would weed the fields by hand. It was hard work, but essential, because weeds compete with rice for light, water, and nutrients. In the fall, he would participate in rice harvesting, threshing, and shouldering the grains to town to sell them.
Kenong recalls the one terrible exception to his contented childhood (see also Video 2):
“When I was 6 or 7 years old, a flash flood occurred and submerged our fields. Every single rice plant died. The consequence was devastating. Many people in the village starved.”
When Kenong was a high-school student, universities that had been closed during the Cultural Revolution were reopened. He took advantage of this opportunity to attend Anhui Agricultural University and study agricultural production. In 1994, Kenong came to the United States to work as a graduate student in the laboratory of David Mackill at the University of California, Davis, to study the genetic basis of rice resilience to flooding. Kenong, Dave Mackill, and I crossed paths when we decided to collaborate on this project, which is featured in this Narrative.
I was excited to be involved in such an important project. I was initially drawn to biology because I loved the natural world, drew joy from being in the mountains, and marveled at the diversity of plants. As a child of a father who was a WW II refugee from Europe and of a mother who grew up during the depression, I had long been aware of the devastation and human displacement wreaked by war and hunger. Although graduate school offered me the chance to explore fundamental mechanisms in plant biology, which I loved, I hoped to do more to help farmers who were struggling to grow food. For this reason, in 1990, I shifted my research focus to rice because it feeds half the world’s people. It was possible, I thought, that one day my scientific research could help poor farmers and their families. This dream was realized in the work with Kenong and Dave and an international team of collaborators in the Philippines, Bangladesh, and India, which many years later would benefit millions of rice farmers.
In this Narrative, the Journey to Discovery features a scientific quest to make a new rice crop that is tolerant of flooding. This is a story of how knowledge of basic plant genetics can be used to benefit farmers. In particular, the Journey features how scientists hunt for genes– in this case, a specific gene that allows rice to tolerate prolonged submergence in flooded fields. Although the story here is about plants, the method that we used is similar to other “gene hunts,” for example, finding genes involved in human diseases.
In the Knowledge Overview, I will step back and provide a broader view of the tools of plant genetics, starting with “early plant breeders” who domesticated the first crops 10,000 years ago to the latest developments in genome engineering. I also hope to demystify the term “genetically modified organism (GMO),” which is widely used but ill-defined and the label “bioengineered,” which will appear on some U.S. foods beginning in 2020.
In the Frontier section, I will describe another more recent scientific story that uses genetic approaches to create rice that produces vitamin A, the so-called “Golden Rice.” This case also exemplifies how modern genetic approaches can be used to fortify foods, which can help diminish malnutrition in many parts of the word.
Finally, I will close with my perspective on the role of science in sustainable agriculture. I am married to an organic farmer who employs diverse farming practices. He has the same goal as I do in my work as a geneticist: we want to help nourish the world, whose population is projected to grow to >11.2 billion people from the current 7.6 billion in the next 80 years, without further destroying the environment. I believe this is the greatest challenge of our time.
Part I: Journey to Discovery —
The Search for a Rice Gene that Controls Tolerance to Submergence
The Problem
Although rice grows well in standing water, most varieties of rice will die if they’re completely submerged for more than 3 days. Each year, 25% of the global rice croplands are inundated by flash floods, which are unpredictable and can occur several times a year. In Bangladesh during the monsoon, roads become waterways for homemade sailboats rigged with cloth, jute, and bamboo, and rice fields are flooded.

Floods cause yield losses ranging from 10% to total destruction. In Bangladesh and India alone, 4 million tons of rice, enough to feed 30 million people, are lost to floods each year (Video 3). These losses disproportionately affect the poorest farmers in the world, where 70 million people live on less than $1 a day.
Explorer’s Question: How might climate change affect the ability of farmers to produce food in Bangladesh?
Answer: The predicted effects of climate change are expected to make rice production even more difficult in the coming years. As sea level rises and glaciers in the Himalayas melt, river systems will experience shorter and more intense seasonal flows, leading to increased flooding of low-lying croplands. Most of the coastal rice production areas in the tropics and subtropics, especially low-lying deltas along the coastlines of South, East, and Southeast Asia, are vulnerable. These areas include the Mekong and Red River Deltas of Vietnam, the Ayeyerwaddy delta of Myanmar, and the Ganges-Brahmaputra delta of Bangladesh. These deltas provide between 34% and 70% of the total rice production in these countries, and any reduction in rice production due to increases in the frequency of flooding will have serious consequences on food security. In Bangladesh, Eastern India, Vietnam, and Myanmar, people get about two-thirds of their total calories from rice.
How does one go about making rice tolerant to flooding? Is this even possible? With such a big goal, it is important to keep in mind that one cannot get there in one big step. You need to find a place to start and let the process of science take you on a journey where step-by-step you develop a better understanding of the biology that enables you to reach your ultimate goal. That process is also exciting because you learn many unexpected things along the way.
For me, this Journey to Discovery began one wintry day in 1996 when Dave Mackill stopped by my office to propose a new collaboration. Dave and Kenong Xu were studying an ancient rice variety that had an amazing property: it could survive two weeks of complete submergence—a situation that often occurs after flooding. Dave asked if I would help them isolate the gene that is responsible for the submergence tolerance trait. My laboratory had expertise in isolating agronomically important rice genes based on their position on the chromosome, and I said yes without hesitation. With our combined expertise, we hoped that we could use genetics to find the gene(s) involved in submergence tolerance. If we were successful, we could introduce the gene into commercial rice varieties grown by farmers in South and Southeast Asia. Then, if we were lucky, we could potentially help millions of farmers produce rice even when their fields were flooded.
This Journey to Discovery takes you through the discovery of a gene that helps to protect rice against prolonged submergence and how this discovery translated into new, flood-tolerant rice crops.
In our hunt for the gene, two clues helped us to get started.
Clues
Clue 1: Identification of a variety of rice that is tolerant to flooding
Plant breeders, using conventional breeding, have had a long track record of success in introducing important agronomic traits (e.g., disease resistance) from wild species or landraces (a domesticated, locally adapted, traditional variety of a species of animal or plant) into cultivated varieties. The International Rice Research Institute (IRRI) is an important modern science-based organization to improve rice production and farming; this organization is based in the Philippines and is dedicated to its mission “to improve the quality of life of those who depend on the rice sector—from the ultra-poor to those moving up the development ladder—and the environmental sustainability of rice production systems” (https://www.irri.org/about-us). Scientists at the IRRI had developed an extremely important resource to begin tackling the challenge of generating a rice variety that confers submergence tolerance to rice: a collection of ~130,000 rice types and wild relatives representing vast genetic diversity. Farmers, consumers, and scientists from countries from all over the world send rice seeds to the IRRI for preservation and sharing.
IRRI scientists decided to test some of the rice varieties in this collection for their ability to withstand prolonged submergence. Most of the rice varieties died if completely submerged for more than a few days. However, scientists at IRRI identified a few rice types that could survive prolonged submergence.
Of particular importance was an ancient Indian rice variety called FR13A (flood resistance 13A), which was unusual in its ability to endure complete submergence for over 14 days. Instead of elongating rapidly to keep leaves above the water so that the plant could continue to photosynthesize (light and gas exchange in unsubmerged tissue is required for photosynthesis), the shoots of FR13A did not elongate until the flood had subsided. This waiting mechanism allowed FR13A plants to conserve energy until conditions were good for reinitiating growth. Thus, the FR13A appeared to activate a “hold-your-breath under water strategy,” which conserves energy reserves until the flood subsides.
FR13A had been known to farmers in Orissa, India, since the 1950s. However, because the quantity and quality of FR13A rice grains are poor, FR13A was not adopted for farm production. For over forty years, breeders at IRRI had tried to use FR13A as a donor parent to introduce the submergence tolerance trait into rice varieties adapted to the local environment that were favored by rice farmers. However, the resulting crosses produced rice varieties that were flood tolerant but lacked many of the traits desired by farmers such as good yield and quality grain. As a result, these rice varieties were also not adopted by farmers.
Explorer’s Question: Why did this conventional breeding method fail to produce a successful flood resistant rice crop?
Answer: Conventional breeding describes an assortment of older methods of genetic improvement, including cross-pollination, hybridization, and grafting (see the Knowledge Overview). In these cases, breeders make genetic changes without knowledge of the precise nature of those changes. In conventional breeding, large sets of genes are combined, many of which are uncharacterized. Because conventional breeding is carried out without knowledge of the exact genes needed to confer submergence tolerance, the breeders unknowingly dragged in undesirable traits from FR13A along with the submergence tolerance trait that they wanted.
Explorer’s Question: Could one use conventional breeding to introduce the submergence tolerance trait from FR13A into other crops such as wheat?
Answer: No. With conventional breeding, gene transfer is typically limited to closely related species. However, there are notable exceptions, such as a cross between rye and barley, which was developed in laboratories in the late 19th century. The resulting hybrid, called “Triticale,” is still sold in some health food stores and has potential as a feed for animals.
Clue 2: Submergence tolerance might be controlled by one or a few genes
A particular trait (a characteristic of an organism) might be controlled by a single gene or by many genes (see the Narrative on Heredity by Tilghman). As an example, in humans, freckle is a trait conferred by a single gene, whereas intelligence is governed by many. If a trait is conferred by a single gene, it is much easier for scientists to study and transfer between plant varieties.
How complex genetically was the submergence tolerance trait of FR13A? Was it the result of one gene or many? Early work suggested perhaps many, which could have made it intractable to study. However, Kenong and Dave as well as other laboratories obtained results indicating that a single chromosome (chromosome 9) and indeed a single region in that chromosome could confer the submergence tolerance phenotype. This was good news, since it suggested that the genetics of submergence tolerance was relatively simple. And if simple (one or a few genes), then we might be able find those genes and transfer them from FR13A into rice varieties that were commonly used in agriculture without transferring other genes that produced unwanted effects on rice production, which complicate conventional breeding strategies.
Explorer’s Question: Was it a waste of money for several laboratories to discover that submergence tolerance was localized to chromosome 9?
Answer: The fact that these experiments were reproduced in several different laboratories was not a waste; rather, it was particularly important in our decision to pursue the time-consuming work of identifying the gene. In biology, environmental variation has a strong effect on results. For example, it is possible that the observed submergence tolerance was not even due to a gene or set of genes but due to factors that were not well controlled. For example, most rice plants can survive some amount of submergence. So if a scientist did not carefully reproduce the conditions in each experiment, s/he might have been misled. One of the difficulties in identifying genetic factors that affect the plants’ resilience to environmental stress is that it can be challenging to control for the environmental variation. Furthermore, science that stands the test time can be reproduced in different laboratories. Thus, these experimental results were important because they indicated that genetic variation, not environmental variation, was the key determinant of submergence. They also provided confidence that a gene(s) might be possible to find.
The Discovery: The hunt for the Sub1 gene
The discovery that the submergence tolerant trait was localized to chromosome 9 suggested that it might be possible to isolate the DNA carrying the gene encoding this trait. With a new grant from the USDA, Dave and I were able to continue to employ Kenong, who had just completed his PhD, as a postdoc to pinpoint the location of Sub1 on chromosome 9. This was a daunting task, especially since the sequence of the rice genome was not known at the time. Chromosome 9 has 22 million DNA base pairs and typical gene is only 3000 base pairs. How does one find the needle in this haystack?
The answer is to make genomic “maps” that can be used to pinpoint the location and then identify the sequence of the gene (Figure 2). The strategy that we used (described later) was to make “maps” of chromosome 9 of increasing resolution to find the gene. We began with a “genetic map” to find the approximate location of the gene on chromosome 9. In a map analogy, the genetic map is analogous to a crude “tourist map” of a city, where the relative locations are correct but not necessarily exactly to scale. After getting to the right neighborhood, one then needs to zero in with great accuracy. This is the “physical map,” where the precise DNA sequence in the location of the gene can be read. This is analogous to the global positioning system (GPS) map of the city, where locations are specified within an accuracy of a few meters.
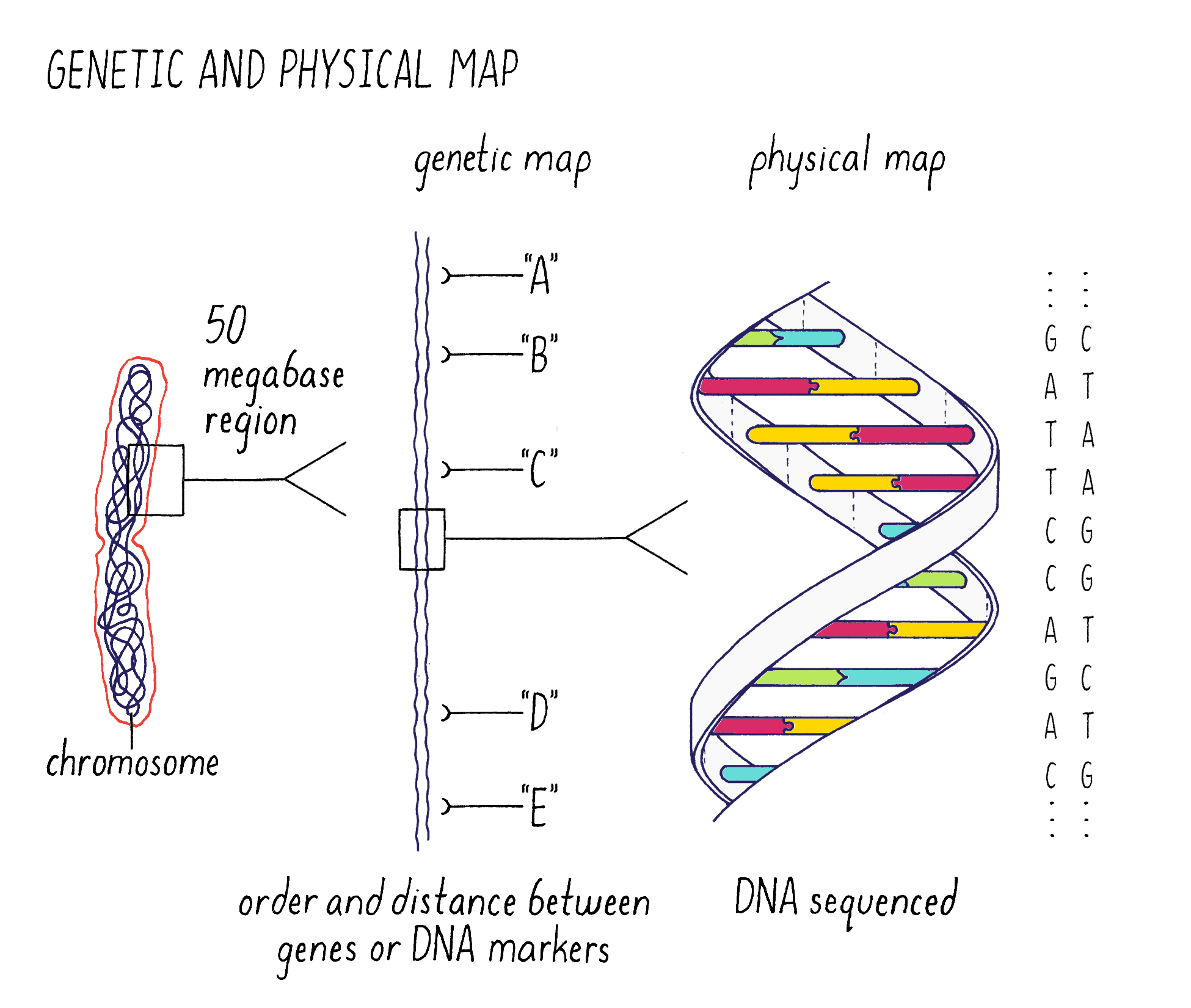
Next, I will discuss the genetic and physical maps in greater detail and illustrate how we used them to pinpoint the gene responsible for submergence tolerance. You will need to understand the terms “gene, allele, chromosome, homologous chromosome, and chromatid” and their relationships to one another. They can be reviewed in Video 4.
Genetic Maps and Gene Linkage
Gregor Mendel formulated the first laws of heredity based upon his studies of various traits of peas (e.g., seed color, texture; see the Narrative on Heredity by Tilghman). In this study, published in 1865, Mendel postulated that traits were specified by “units of heredity” (now known as “genes”) and that these units segregate randomly from one another when they are passed from parents to offspring. While true for the particular traits that Mendel studied, later studies by Thomas Hunt Morgan, beginning in 1910, found that independent and random segregation of traits is not always true.
When Morgan and student Alfred Sturtevant, who was working with him, studied the laws of inheritance in fruit flies, he observed that certain traits were “linked” to particular chromosomes. He first discovered this by showing that a gene for eye color followed the behavior of segregation of the sex-linked “X” chromosomes in the fly. This led to a general conclusion that genes (and the traits that they encode) found on different chromosomes would be expected to segregate independently because of how chromosomes behave during the process of meiosis. If unfamiliar with the basics of meiosis, please see this short Whiteboard video (Video 5).
Let’s investigate what would happen if traits encoded by two distinct genes (A and B) are on the same or different chromosome.
If the two genes are on different chromosomes (Figure 3), the traits associated with those genes would segregate independently of one another because of the random ways in which the two homologous chromosomes can pair with one another during meiosis I. In the example of Mendel’s peas, the traits for pea seed color (green or yellow) and pea seed texture (wrinkled or smooth) segregate independent of one another (see the Journey to Discovery in the Narrative on Heredity by Tilghman).
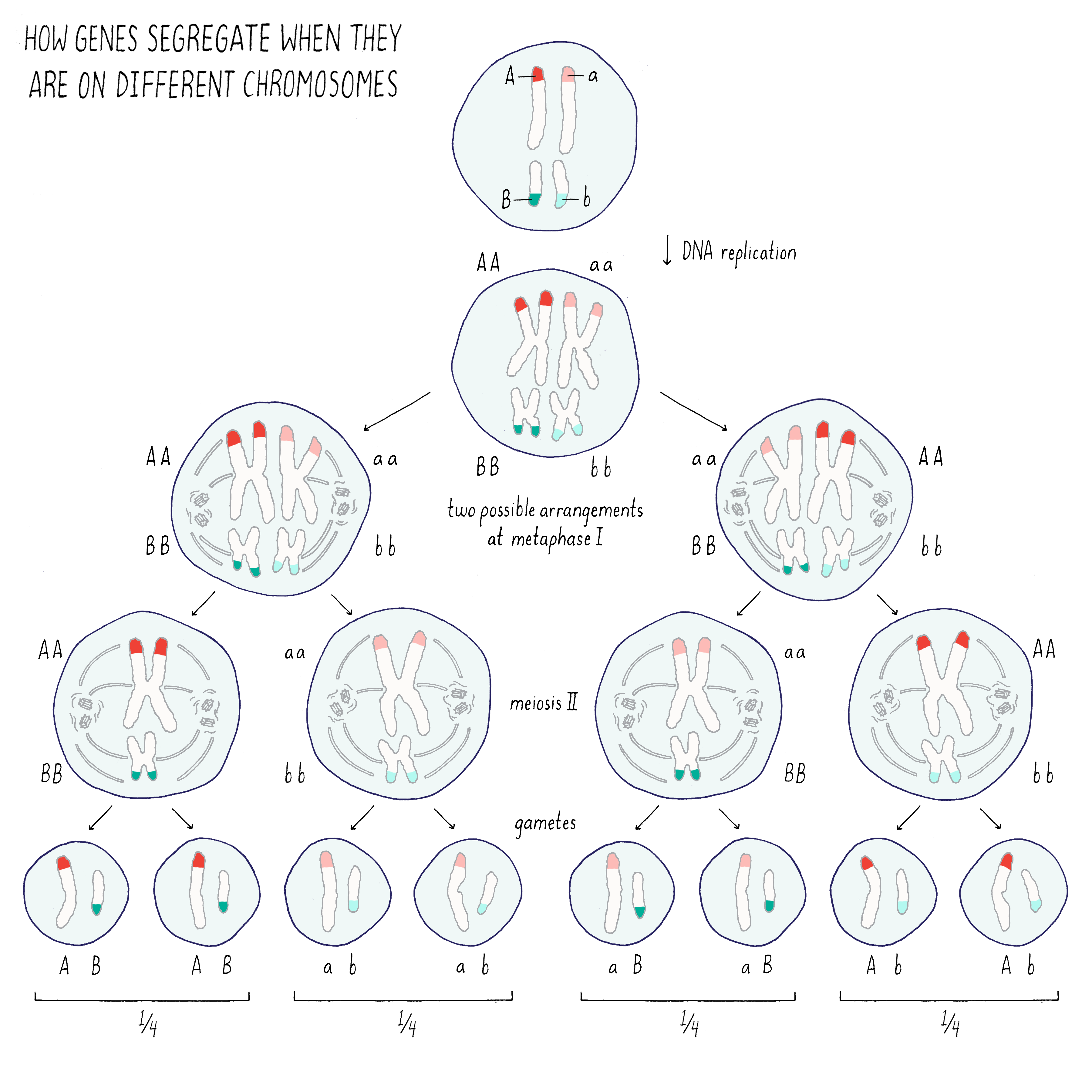
Explorer’s Question: From the drawing above, what would you expect to happen for the two traits if the genes A and B were on the same rather than different chromosomes?
Answer: In this case, the genes A and B (and their associated traits) would segregate together during the process of meiosis.
However, there is an additional wrinkle to the mechanism of gene segregation – homologous chromosomes can swap segments of DNA through a process called either “crossing-over” or “recombination.” This process of recombination occurs during the first stage of meiosis when the homologous chromosomes pair with one another(Figure 4).
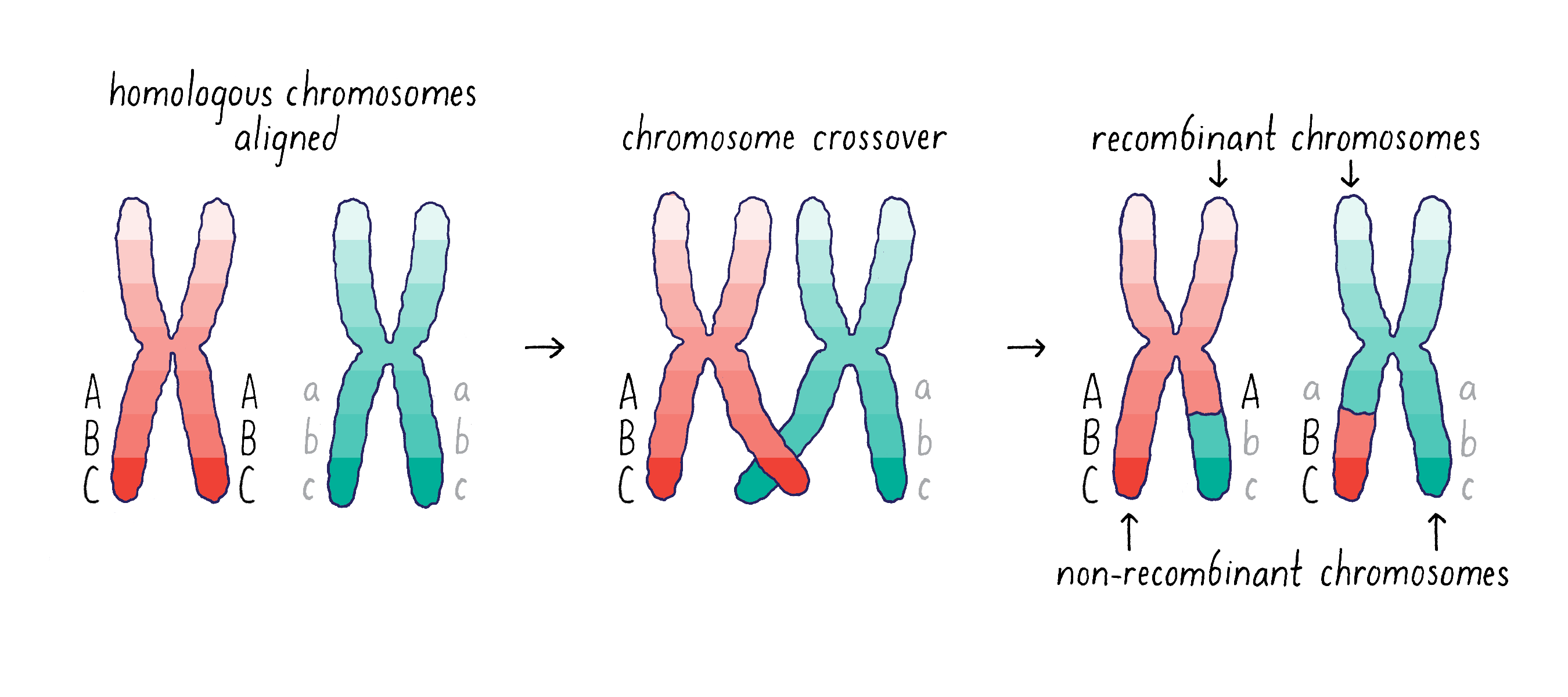
Morgan and his student Alfred Sturtevant came to the important conclusion that recombination provides a means of mapping the distance between genes on a chromosome (see Video 6). If two genes are separated further apart (for example, Aa and Cc in Figure 4), then it is more likely that a recombination event will occur between them. This recombination will mix parental and maternal alleles on the recombinant chromosome (Abc and aBC). However, if the two genes are close (for example, B and C), then it will be less likely that a recombination event will occur between them. Thus, the big insight of Sturtevant and Morgan was that by measuring the frequency of recombination, they could determine the linear order of the genes on a chromosome and obtain information on the distance between any two genes. This genetic distance is measured in “map units,” where 1 map unit is defined as the interval in which 1% crossing over takes place. The map unit is sometimes called centimorgan (cM) in honor of Morgan.
Making a genetic map to localize the submergence tolerance genes
To narrow down the region on chromosome 9 that contains the Sub1 gene, we first had to create a high-resolution map. To do so, we used known DNA markers on chromosome 9 (similar to that shown in Figure 2). A DNA marker is a DNA sequence that varies among individuals (having arisen due to a mutation at some point in the past within a species). Variations in genes are frequently referred to as “alleles”. Scientists can use markers to distinguish the DNA between parents. By analyzing the DNA markers of the offspring, one can determine whether a specific region of a chromosome was inherited from either the mother or the father.
In addition to reading the text, you can also hear Kenong tell his story of the research involved in finding the Sub1 gene, which took him 10 years from start to finish (Video 7).
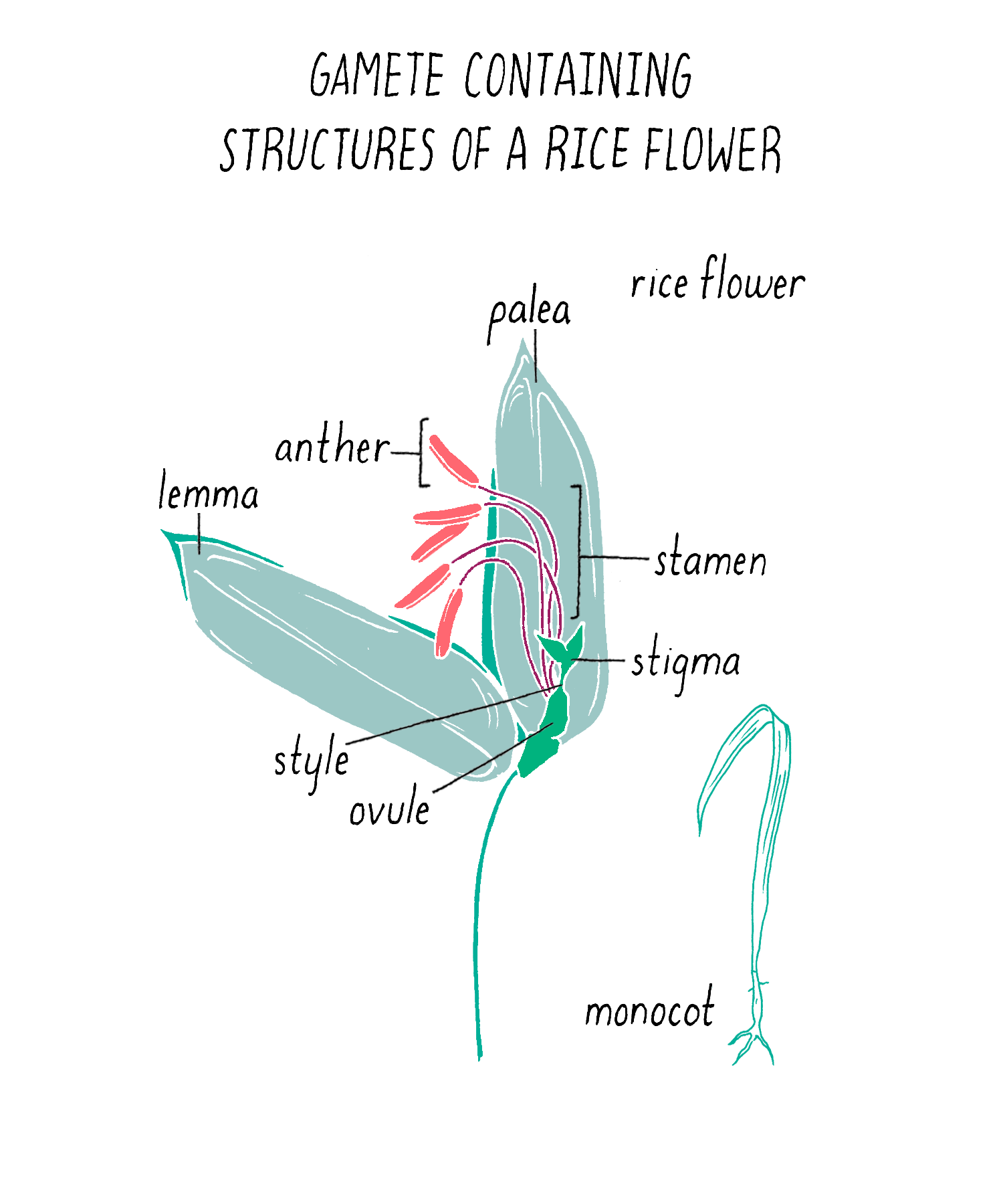
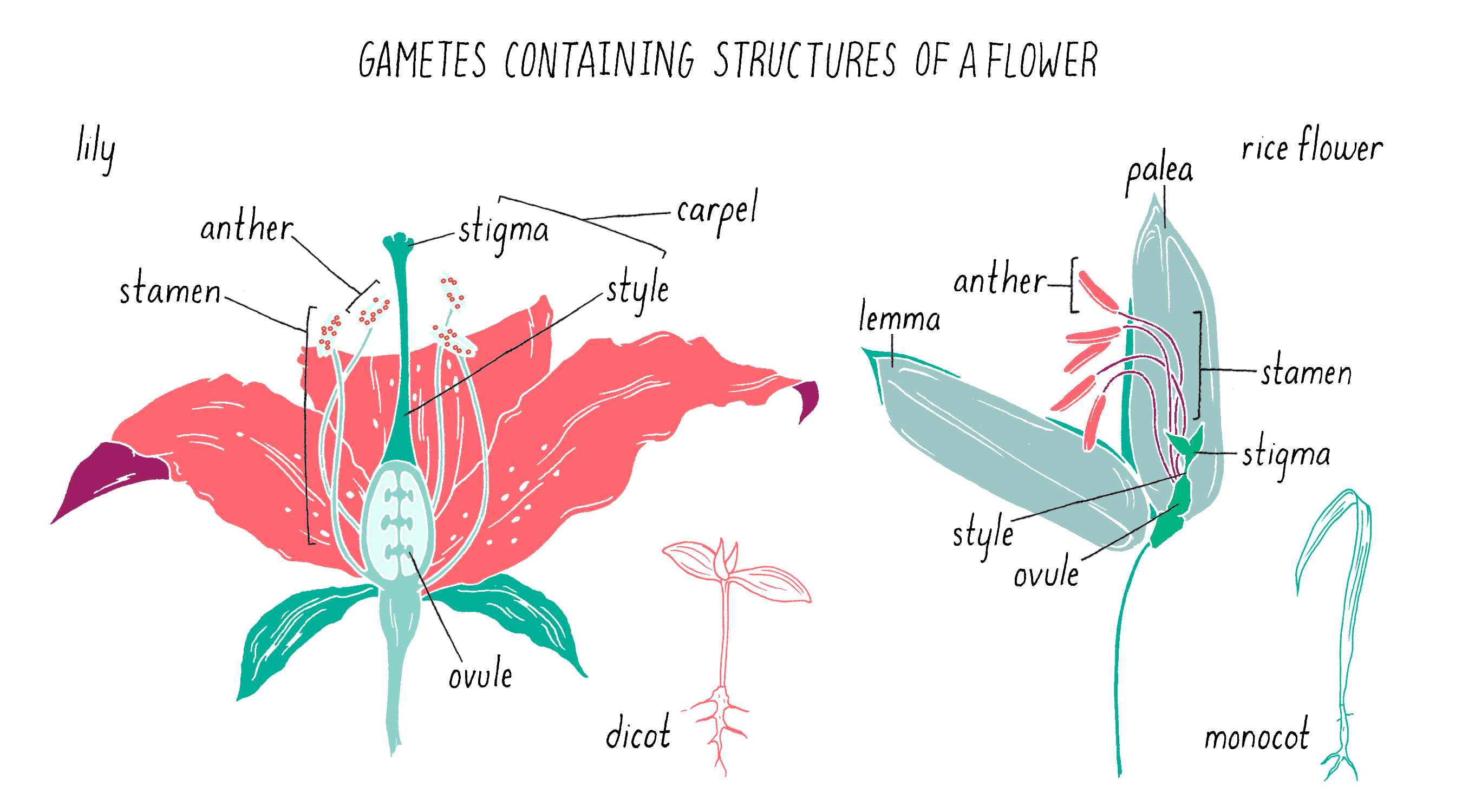
To initiate the mapping, Dave and Kenong set up crosses between two varieties of rice: one that was submergence-sensitive and another that was submergence-tolerant. For this experiment, Kenong grew each plant until the rice flowers emerged (Figure 5). Then, he cut off the top half of the bracts (lemma and palea) with scissors to expose the anthers (male flowering part that produces the pollen) and stigma (female part that carries the eggs) inside (Figure 5). He then used tweezers to carefully remove the anthers from each flower without harming the stigma. Once the anthers were removed, he moved this “emasculated” plant close to the other plant. He placed bags over the two flowers so they can mate. Every few days, at mid-day, he tapped the bag so the pollen from one parent would fall onto the stigma of the other parent. After three days, the pollination was complete and the bags were removed. One week later, the seeds began to form.
The first cross between the submergence-tolerant plant (traits symbolized by green color and short stature, respectively) and the farmer’s locally adapted, but submergence-intolerant, plant (traits symbolized by brown color and tall stature) is shown in Figure 6. These “F1” plants carried one chromosome 9 from the tolerant parent and one chromosome 9 from the intolerant parent (Figure 6; see the Narrative on Laws of Inheritance by Tilghman).
The F1 plants can now be self-fertilized (male and female gametes combined from the same plant) and give rise to F2 generation plants. These F2 plants can inherit either two chromosomes from the tolerant parent (double green; homozygous), two chromosomes from the intolerant parent (double brown, homozygous), or one from each (green-brown, heterozygous) (Figure 6; see the Narrative on Laws of Inheritance by Tilghman). But occasionally, the gametes of the F1 plant may have undergone a crossover event as shown in Figure 4. That “recombinant event” would create a chromosome with a region from tolerant plant and intolerant plant (brown-green chromosome) which will get passed down to the F2 generation. These are the recombinants, an example of one of many possibilities shown in Figure 6. Some of those recombination events could introduce the region with the Sub1 gene into the chromosome from the intolerant plant, now allowing that chromosome to convey a phenotype of submergence tolerance.
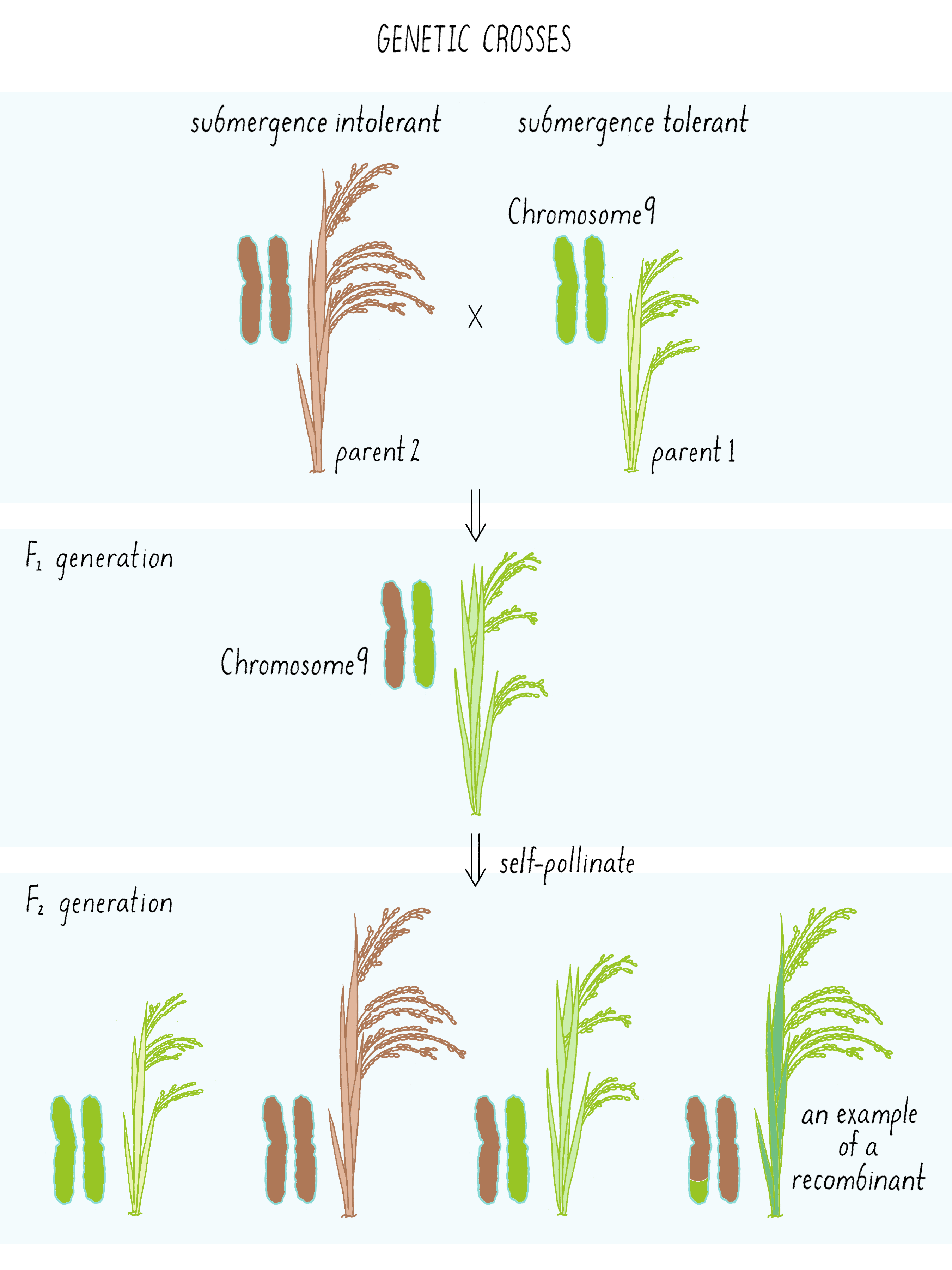
But how does one tell which chromosome is from which plant? And how does one tell if there is a recombination event and where the break point might be in the chromosome. Since the chromosomes are not neatly colored as in Figure 6, one needs another signature and that signature comes from the DNA sequence. The DNA sequences in the chromosomes from the tolerant and intolerant strains are very similar but not identical. These subtle differences can be detected and used as “markers” from each chromosome. In Figure 7, we show DNA markers in the approximate vicinity of the Sub1 gene. We will label the DNA markers from the tolerant plant as A, B, C, and D and from the intolerant plant as a, b, c, and d.
The initial work indicated that the Sub1 gene was somewhere between A and D. But was it between A-B, B-C, or C-D?
A further mapping experiment had to be done to further pinpoint the gene location, which involved looking for recombination events in the A-D region. However, since the A-D region is a relatively small region, the recombination would be expected to be rare, as Morgan and Sturtevant described. To search for them, Kenong extracted DNA from thousands of F2 plants and tested for the presence of markers that would indicate a crossover event between A and D. In addition, he also had to determine whether the Sub1 gene was present in that recombinant chromosome. Since the Sub1 DNA sequence was unknown (that was the goal), we had to test for its presence by analyzing for the submergence tolerance phenotype. Knowing whether that recombinant chromosome conferred the submergence tolerance phenotype required two more steps, self-fertilization of the F2 plants to generate F3 offspring and then testing these F3 for submergence tolerance properties, which you can read about in Dig Deeper 1. Here, we just describe the analysis of 3 recombinant plants that further pinpointed the location of the gene.
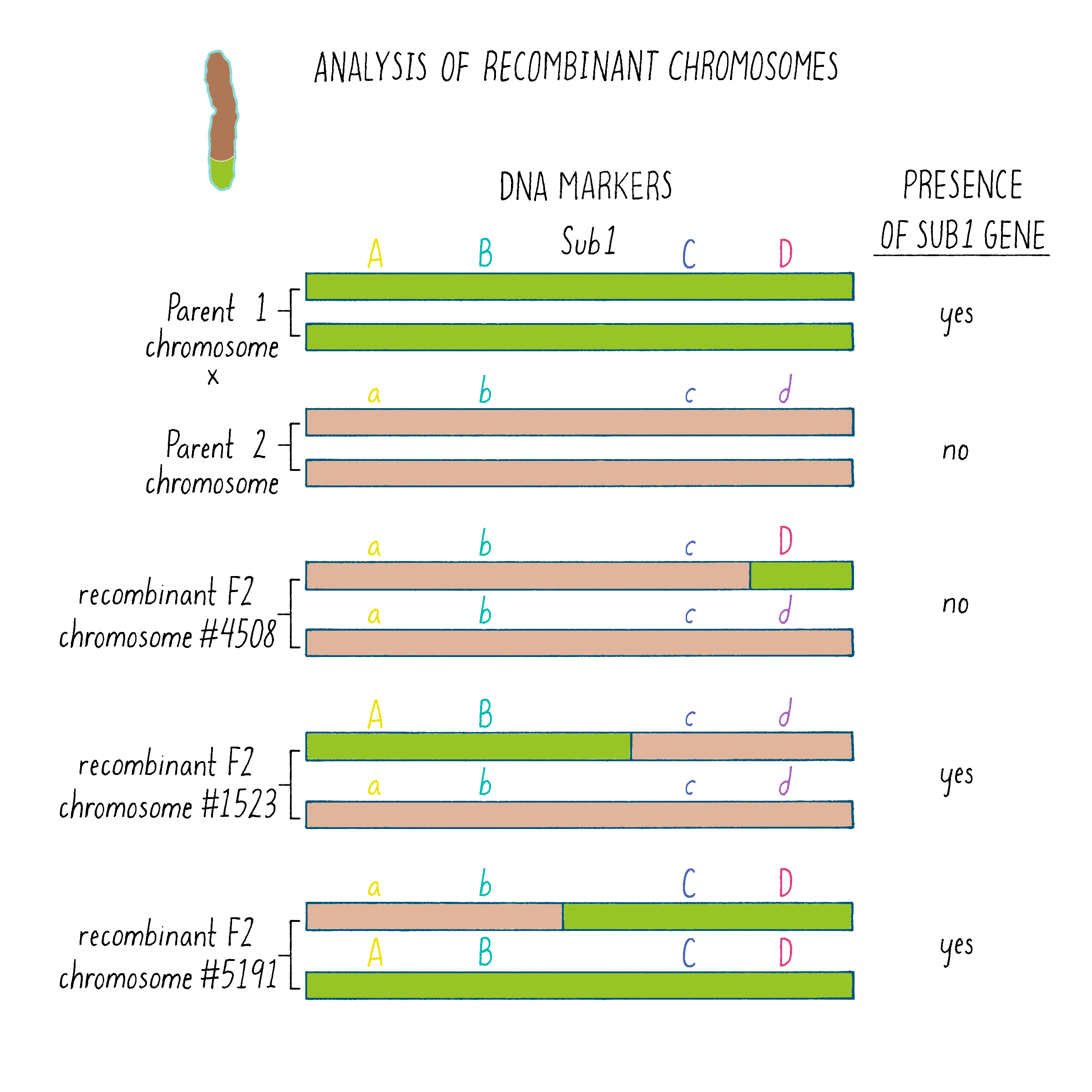
As shown in Figure 7, in the plant #4508, the recombinant chromosome had markers a b c D (Figure 7). This chromosome was unable to confer tolerance (i.e., did not contain the Sub1 gene) even though it contained the D region from chromosome 9 of the tolerant plant. Thus, the Sub1 tolerance gene could not be at or to the right of the “D” region.
In plant #1523, the recombinant chromosome had the markers A B c d and conferred tolerance (Figure 7). The recombination that brought in A B markers from chromosome 9 of the tolerant plant (shown in green) must have brought in the Sub1 tolerance gene. The gene must be located to the left of the “C” region.
In plant #5191, the recombinant chromosome had the markers a b C D and also conferred tolerance (Figure 7). Therefore, the recombination that brought in the C D markers from chromosome 9 of the tolerant plant must contain the Sub1 tolerance gene. That gene must be located to the right of the B region.
Since recombinant chromosomes in both 1523 and 5191 have the Sub1 tolerance gene, the gene must lie in the region that they share in common from chromosome 9 of the tolerant plant (overlapping green regions in Figure 7). This shared region lies between markers B and C. The B-C region still contains 182,000 nucleotides (182 kb), which seems like a lot, but it is a 2500-fold reduction in size compared to the total rice genome (450 million base pairs). But 182 kb is still a large region, and thus, we had more work ahead to find the gene.
Making a physical map to localize the Sub1 gene
Once we had defined the genetic map, our next step was to assemble a “physical map.” Now, rather than discussing map distances in centimorgans, phenotypes, or DNA markers, here we are discussing physical pieces of DNA and DNA sequences. At the time we started the project, the rice genome had not yet been sequenced so the DNA sequence of chromosome 9 was mostly uncharted territory.
The first step in this process was to create “libraries” of DNA clones from submergence-tolerant rice that carry pieces of DNA (Figure 8). To do this, we cut the genome into smaller pieces with restriction enzymes and insert these pieces into plasmids that can replicate in bacteria.
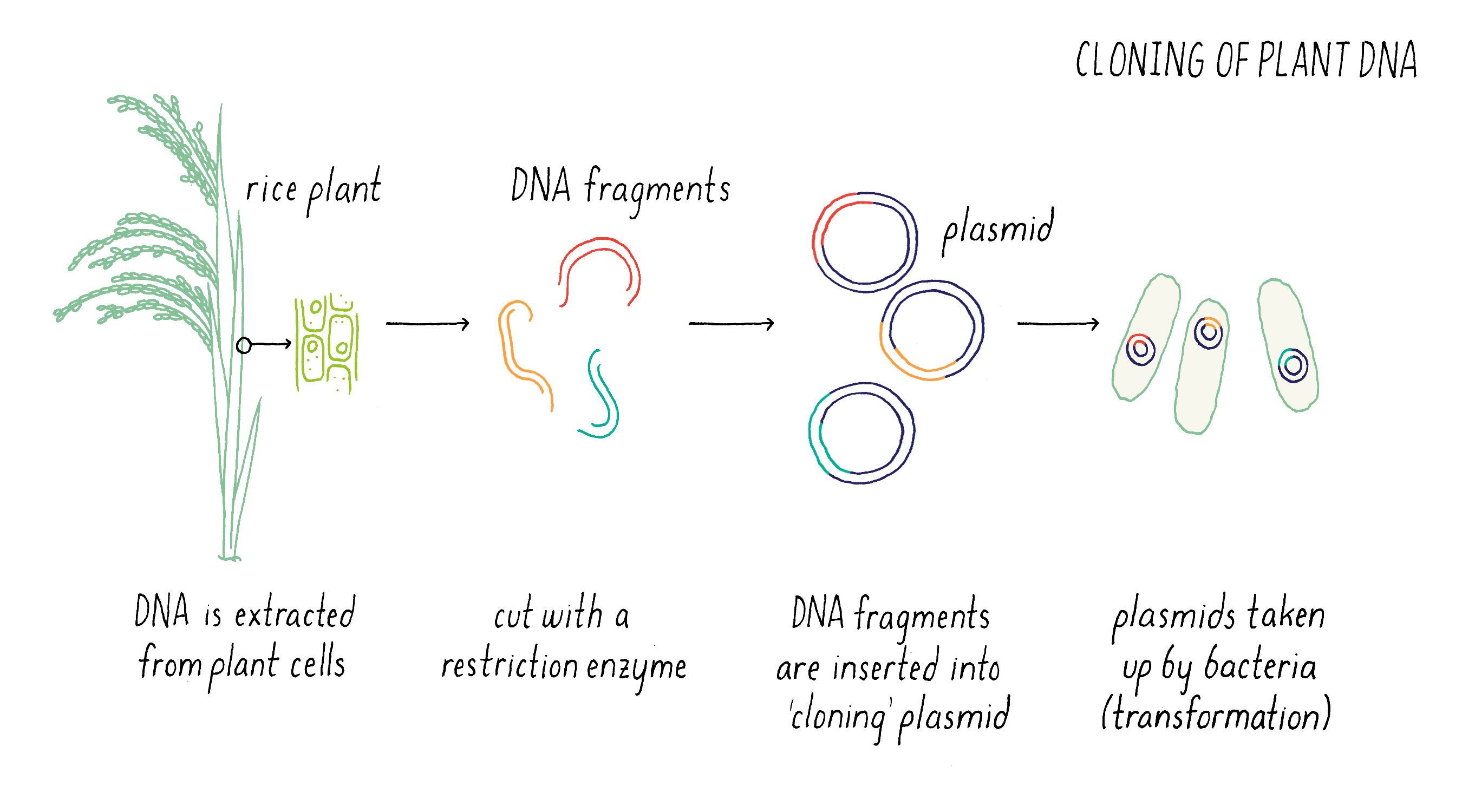
We then identified those clones that contained fragments of DNA that contained the DNA markers B and C (see Figure 7) by hybridization. Some clones contained the DNA sequence of marker B but not marker C, while other contained the sequence of marker C but not marker B (Figure 9). However, there were regions of the cloned sequences that overlapped with one another; in other words, the nucleotides sequence of 3′ end of one clone containing marker B overlapped with the 5′ end of another clone containing marker C. In this manner, we were able to stitch together (on a computer) the complete and contiguous DNA sequence between markers B and C (if you want to learn more, see the Whiteboard Video on Bioinformatics).
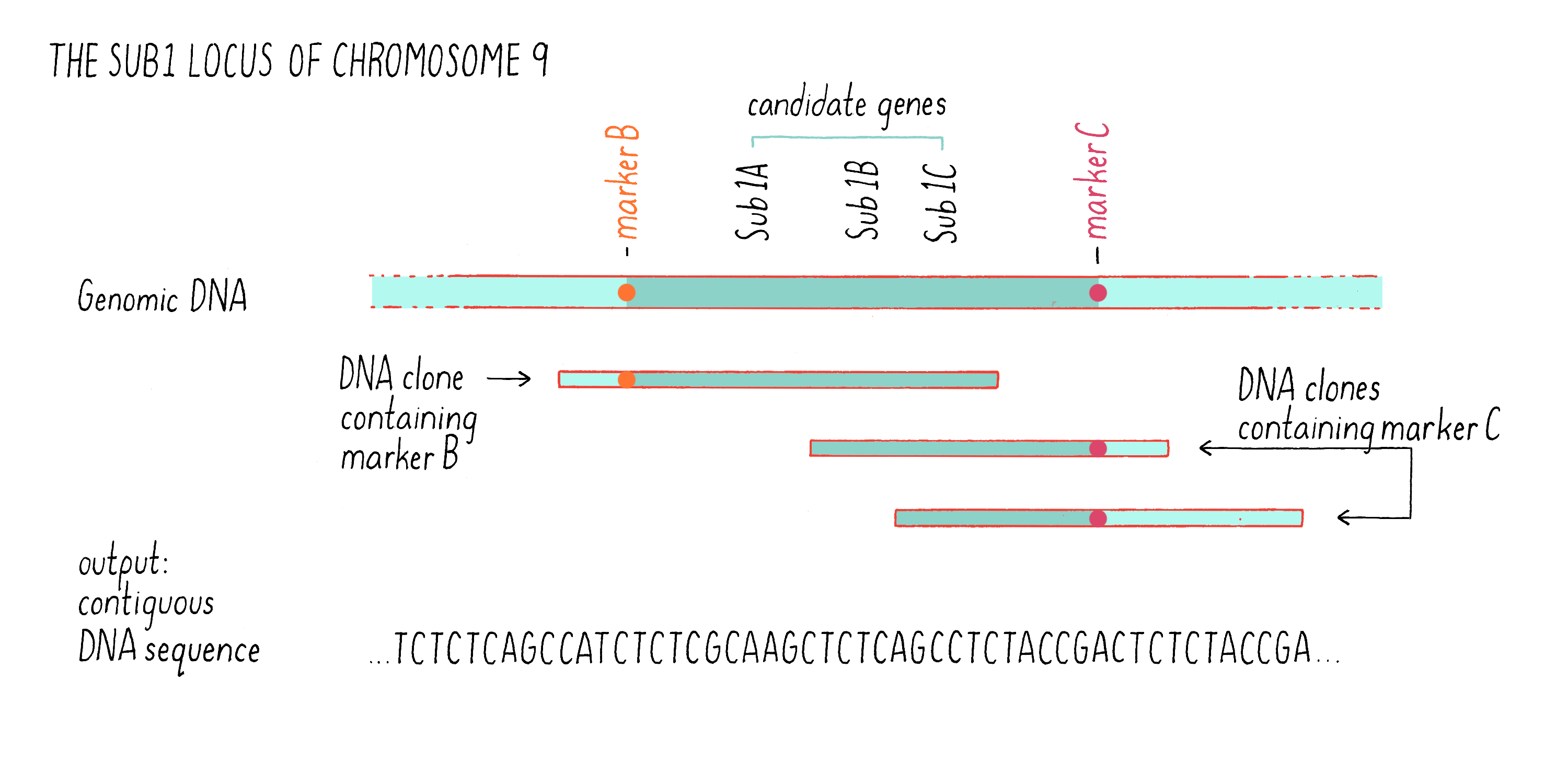
With the complete DNA sequence between markers B and C in hand, we then searched for sequences that are characteristic of genes, most specifically looking for sequences that carried a typical gene promoter, followed by a start codon and then by a stop codon. One day, after months of DNA sequencing and analysis, Kenong came in my office with a smile. He showed me that he had identified 13 genes in the 182 kb Sub1 genetic region between markers B and C (a region of a chromosome is also sometimes called a “locus”). We were excited to discover that three of these genes, which we designated Sub1A, Sub1B, and Sub1C (see Figure 9), were predicted to encode proteins that were similar to a class of transcription factors that had been previously characterized as responsive to environmental stress (submergence is a form of environmental stress). These were excellent “candidate genes” for being involved in submergence tolerance (see Dig Deeper 2 about how Julia Bailey-Serres and team showed how Sub1A responds to and regulates hormones to alter plant growth and function during submergence).
But which one of the three Sub1 genes (if any) might be the one that we were looking for? We rationalized that the “correct” gene might reveal itself by responding to submergence stress by turning on or off its transcription. Kenong tested this idea by isolating mRNA from Sub1 rice plants that had been subjected to submergence at different time points. Using a technique called “Northern blot analysis” (Dig Deeper 3), he could measure the amount of mRNA of each Sub1 gene before and after flooding. Kenong noticed that only one of these genes, Sub1A, was “switched on” rapidly (even after 2 hours) after submergence and began to transcribe mRNA (Figure 10; Dig Deeper 3).
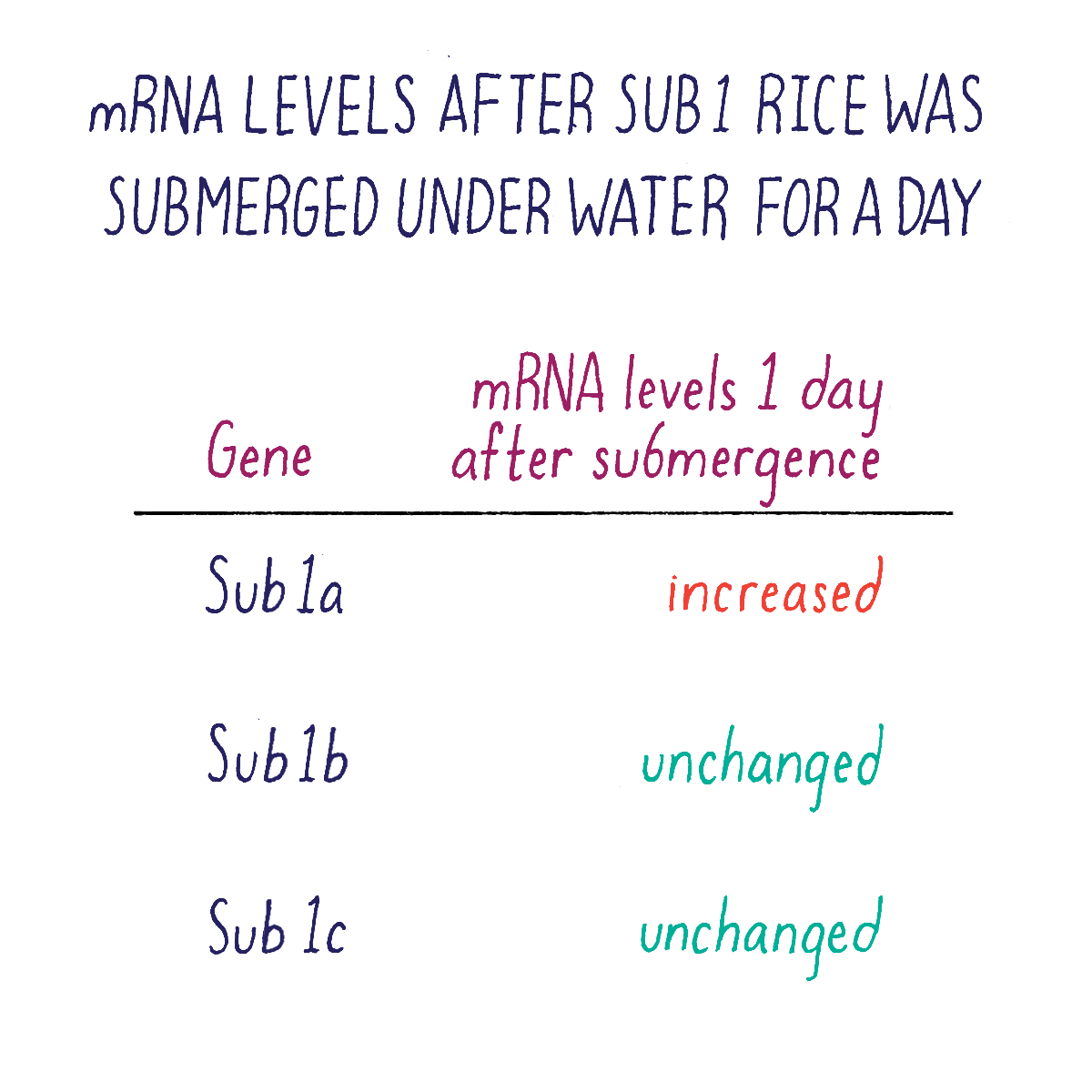
Kenong next acquired DNA samples from rice varieties selected by researchers at the International Rice Research Institute. His DNA analysis revealed that rice varieties were intolerant to flooding lacked the Sub1A gene, whereas those tolerant to submergence carried the Sub1A gene. Together, these data suggested that Sub1A controlled the submergence-tolerant response. Sub1b and Sub1c were not good candidates because they were present and expressed in all rice varieties, even those that were intolerant of submergence.
Explorer’s Question: The data described above revealed that the Sub1A gene is found in submergence tolerance rice varieties but not found in varieties that are intolerant. Are you convinced at this point that the Sub1A gene alone governs flood tolerance?
Answer: This data is a correlation – i.e., the presence or absence of the Sub1A gene is correlated with the trait of interest. While very strongly suggestive, such evidence alone does not show cause and effect. In other words, the data does not prove that the Sub1A gene is responsible for submergence tolerance. Additional evidence is needed.
Explorer’s Question: What strategies might you take to gain stronger evidence that Sub1A is responsible for submergence tolerance?
Answer: There are two general ways in which scientists associate a gene with a phenotype. One way is not “knock out” the gene and see if the trait goes away. This is a common strategy. However, if there is “redundancy” of multiple genes playing similar roles in conferring a trait, then the gene knockout will not eliminate the phenotype. Another strategy is to see if the gene confers a gain-of-function when transferred to a variety that lacks the trait. We chose the gain-of-function strategy.
Validation of Sub1 gene function
To validate whether the Sub1A gene (name simplified afterwards to Sub1 gene) is truly responsible for protecting plants against flood submergence, we introduced the Sub1 gene into a submergence-sensitive rice that lacked the Sub1 locus (see the Knowledge Overview on how to use bioengineering to introduce a gene into a plant). Our prediction was that if Sub1 encoded the submergence-tolerant trait, then the newly bioengineered plant would be tolerant to submergence.
Next, we harvested seed from the plants carrying Sub1 and tested whether the bioengineered seedlings could survive submergence for 17 days. We found that the control plants lacking Sub1 (shown on the left side of Figure 11) were weak, spindly, and pale and did not recover after submergence ended. In contrast, the rice plants that carried the introduced Sub1 gene survived the extended time under water and recovered well (see the right side of Figure 11). This data was important because it showed that transferring the Sub1 gene to another rice variety was sufficient to confer submergence tolerance.

The result was exciting – our hunt for a gene that confers survival during flooding came to a successful conclusion. However, we still did not know if our laboratory and greenhouse experiments would translate in the field and thus whether Sub1 would be sufficient to help rice farmers. This leads to the next part of the story.
What Happened Next?
During the time that we were identifying the gene at UC Davis, Dave Mackill moved to IRRI where he began a breeding effort to introduce Sub1 into Swarna, a variety grown by millions of rice farmers in India and Bangladesh (Video 8). One strategy was to introduce the Sub1 gene directly into Swarna using bioengineering (see the Knowledge Overview for more details). This is effectively what we did in our research study at UC Davis to demonstrate that Sub1 confers submergence resistance in the experiments I described above. However, if bioengineered, the rice would be subject to a number of complex regulatory controls. Furthermore, regulations regarding the planting of bioengineered rice were not yet established in the countries where it was needed most (Southeast Asia), and it could have taken many years before a bioengineered Swarna rice variety was approved for planting by the appropriate regulatory agencies.
However, because Sub1 is a rice gene, bioengineering was not necessary to introduce the gene. Instead, it could be introduced from one rice variety to another through “marker-assisted breeding” (see the Knowledge Overview) between a rice variety containing the Sub1 gene and the Swarna variety. This approach uses DNA markers to screen through many plant progeny that carry Sub1 but that don’t carry genes that could negatively affect desirable traits. Plants developed through marker-assisted breeding, which uses DNA technology to select the best offspring obtained by conventional breeding, are not subject to regulation. Dave and his team used the marker-assisted breeding approach to introduce the Sub1 gene into Swarna. He then carried out several years of field tests in collaborations with rice breeders in India and Bangladesh.
The researchers found that the newly developed Swarna-Sub1 varieties could withstand 14 days of submergence in the field, and their yields and tastes are the same as the parent variety. In each of the 9 years from 2008 to 2016, farmers in Bangladesh and India were able to harvest more grain from flooded fields planted with Sub1 varieties compared with fields planted with conventional varieties (1.5 tons/hectare more grain on average). In the absence of submergence, Sub1 plants had no observable differences compared to plants without the Sub1 gene. However, following flooding for >20 days, the normal Swarna rice produce little or no yield of rice, while the Swarna-Sub1 had much higher rates of survival and improved yield of crops (Figure 12).
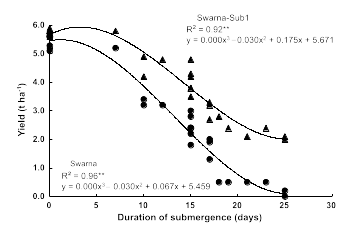
Substantial efforts have been undertaken by IRRI and national programs to incorporate Sub1 into additional local popular rice varieties grown by farmers in Vietnam, India, Bangladesh, and Thailand. In 2017, 6 million farmers grew Sub1 rice, setting a record for the most rapidly adopted rice variety in the history of modern farming. The Sub1 varieties, distributed through government-breeding stations, have enhanced the productivity of the farms of poor families in India, with the highest benefits going to disadvantaged farmers who historically have cultivated plots that are flood prone.
One of the important aspects of the Sub1 project was its highly collaborative nature. Specifically, the combination of molecular geneticists, physiologists, breeders, and farmers working together and freely sharing material greatly enhanced the success of this project. For example, early on, prior to publication, our team shared the Sub1 genes and our Sub1 rice lines with other laboratories. UC Davis did not file a patent on the Sub1A gene because patenting could have hindered rapid deployment of Sub1 varieties throughout Asia. This “open science” approach facilitated collaborations that advanced our understanding of Sub1 and that benefited farmers in an expeditious manner.
Finally, you can hear Kenong’s reaction to their research work achieving a practical outcome of helping rice farmers in Asia (Video 9).
Part II: Knowledge Overview —
Old and new strategies of plant genetics
The history of plant science is among the oldest. Our ancestors ~12,000 years ago began experimenting with plant breeding and artificial selection, giving rise to domesticated plants and early agriculture. Today, all of our principal food crops come from varieties that have resulted from artificial selection. These crop varieties depend on artificial environments (i.e., farms) for their survival. However, the last thirty years have brought new and more precise bioengineering strategies to crop development. In this section, we will compare and contrast the various methods of traditional and modern plant genetics (Figure 13).
Classical plant genetics
Early agriculture and artificial selection
Natural selection, the process by which organisms that are better adapted to their environment have a greater chance of surviving and producing more offspring, has led to the generation of all life forms on Earth over 3.8 billion years. However, the rules changed ~9,000 to 12,000 years ago when humans began to domesticate plants and animals. Now humans, not nature, could choose which organisms have pleasing properties and select those for mating and propagation. Over the millennia, this choosing, known as artificial selection, has resulted in a diverse array of domesticated species that are unlike their ancestors. For example, dogs, unlike their wolf ancestors, have been bred for docile traits, hunting, or appearance.
Similarly, modern food crops are descendants of wild plant species. Approximately 12,000 years ago, farmers began selecting plants with favorable characteristics and replanted the seeds from those plant types. These farmers were indeed early scientists who were exploring and understanding the natural world and beginning to manipulate heredity. Without realizing it, early farmers were selecting for a particular genetic makeup. Today, virtually all foods sold in markets are products of selective breeding by humans ( Video 10).
The plants that evolved through this human artificial selection differ dramatically from the original species. For example, modern corn evolved from a wild grass called teosinte, which has tiny kernels and cobs (see Video 11). Evidence from archaeological and genetic studies suggests that maize was bred and cultivated by early inhabitants of Mexico as early as ten thousand years ago. Scientists have uncovered genetic variants with larger corn cobs, introduced through human selection, in caves of Mexico dating from ~6,000 years ago. Scientists hypothesized that spontaneous mutations gave rise to mutants that retained kernels intact on the cobs or had more rows of kernels, unlike the teosinte ears, which shatter into individual kernels. Our ancestors must have noticed these mutants and then selectively propagated them.
Another example of artificial selection by farmers is the precious sticky rice of Thailand. Sticky rice lacks the starch amylose, which constitutes up to 30% of the total starch in nonsticky rice. The lack of amylose results from a mutation in a gene called Waxy, which encodes an enzyme that synthesizes amylose. Sticky rice is an important culinary and cultural component throughout East Asia and is used in festival foods and desserts. In upland regions of Southeast Asia, it is a staple food in many homes. Ten percent of the rice traded internationally each year is sticky rice.
Where did sticky rice come from? Laotian Buddhist legend places the origin of sticky rice at about 1,100 years ago, although Chinese folklore indicates that it was in existence well before that, around the time of the death of the poet Qu Yuan more than 2,000 years ago. History can be imprecise or contradictory, but the sticky rice itself can provide clues to its origin – if we sequence its DNA.
Rice varieties carry the same set of genes. However, a gene in one variety is often slightly different from its counterpart in a related variety, meaning that its DNA sequence is very close but not identical. The same is true in different humans, that is, the basic nature of genetic diversity. The researchers hypothesized that if a single breeder had developed a really good sticky rice variety and then shared it with other breeders, the new varieties would all contain exactly the same mutation in the Waxy gene. That one mutant plant 1-2000 years ago would be the great, great, … great grandparent of all of the sticky rice today. On the other hand, if sticky rice arose at different times in the history of agriculture, then one would expect to find different mutations for at least some sticky rice varieties, and not the identical DNA signature.
To understand the origin(s) of sticky rice, geneticists looked at the sequence of the Waxy gene in 105 rice varieties. They found that all those that had the sticky texture carried almost the same sequence in their Waxy gene, including the same mutation that knocks out the production of amylose. This result suggests that the mutant sticky rice originated only once, most likely in Southeast Asia. Some observant farmer over one millennium ago liked the adhesive quality conferred by that single Waxy mutation and preserved that particular trait through breeding by incorporating it into new varieties with other desirable traits. The development of sticky rice is a good example of how plant breeders choose plants with a desirable spontaneous mutation (see the Narrative on Mutations by Koshland) and select it for future breeding and maintain selection of the beneficial trait.
Artificial Selection by controlled pollination
Pollination is the transfer of pollen grains (i.e., male gametes) from the stamens (i.e., male reproductive organs) to the plant carpel (i.e., female reproductive organ), which contains the ovule (i.e., female gamete; Figure 14). The receptive part of the carpel is called a stigma in the flowers of angiosperms (i.e., flowering plants). Open-pollinated plants reproduce in one of two ways: self-pollination or cross-pollination between two plants (by wind, insects, or water).
Self-pollination
The pollen is delivered from the male stamen to the female carpel of the same plant. Beans, lettuce, peas, and tomatoes are self-pollinating plants and do not require isolation to remain true to type.
Cross-pollination
A pollinator (e.g., wind, insects, humans) delivers pollen to the female carpel of a different individual of the same species (or a different but closely related species). An example of a cross-pollinator is corn. Corn plants carry the stamens on tassels on top of the plant. The ears, which carry the carpels (that develop into kernels), are produced below on the main stem. The prominent stamens allow the plant to better spread the pollen to ears of other plants. In this way, corn is adapted for cross-pollination. Beets, brassicas, carrots, corn, and squash are cross-pollinating plants and require isolation in the field to keep the varieties true to type.
Controlled pollination
Farmers and scientists learned early on to use pollination to control the breeding in plants. For example, Gregor Mendel in the 19th century, who uncovered the basic principles of inheritance of traits (see the Narrative on Heredity by Tilghman), used selective breeding through controlled pollination. Mendel took great pains to avoid cross-pollination in his experimental garden. His choice of the pea for his experiments was primarily because they are self-pollinating. Going one step further to control the breeding, he removed the stamen from one pea, where the pollen is produced; that plant only would have the female ovule and could not self-pollinate. He then introduced male pollen from another pea plant to that ovule. In this way, he could perform a genetic “cross” between parents with two different traits and could investigate how those traits were passed on to the progeny.
By controlling the crosses between plants, breeders are able to more precisely manipulate plant species to generate desired combinations of traits for specific purposes. In this artificial selection process, the breeder transfers the male pollen grains to the female part of the flower. Effectively the breeder can control what “dad” plant mates with which “mom.” Early artificial selection approaches were often quite crude, resulting in new varieties only through a combination of trial and error, and without knowledge of the precise function of the genes that were being transferred.
Hybridization
Hybrids result from controlled cross-pollination between two distinct plant varieties. Cross-pollination of rice is described above. Although each plant species has a different flower structure, in each case, cross-pollination involves placing the pollen of one plant onto the stigma of another. The hybrid seed that results from this cross carries characteristics that farmers’ value, such as high yield, disease resistance, and good flavor. The seeds produced from hybrid parents segregate for various traits and are not “true to type.” In other words, the progeny do not have the same type of features as the hybrid parents. Thus, the plants in the next generation do not resemble the parent plants. For this reason, growers who prefer hybrids buy seeds every year from seed companies. Hybridization was developed in the 1920s, long before the advent of bioengineering.
Grafting
Grafting (Figure 15) brings two different species physically together. The upper part of one plant (scion) is combined with the lower part (rootstock) of another plant. For example, in California, commercial walnut trees are grafts. They carry a rootstock from a native species of walnut, which is tolerant of root diseases and the scion from an English walnut species that produces the nuts. In this case, the seeds and offspring remain English walnut.
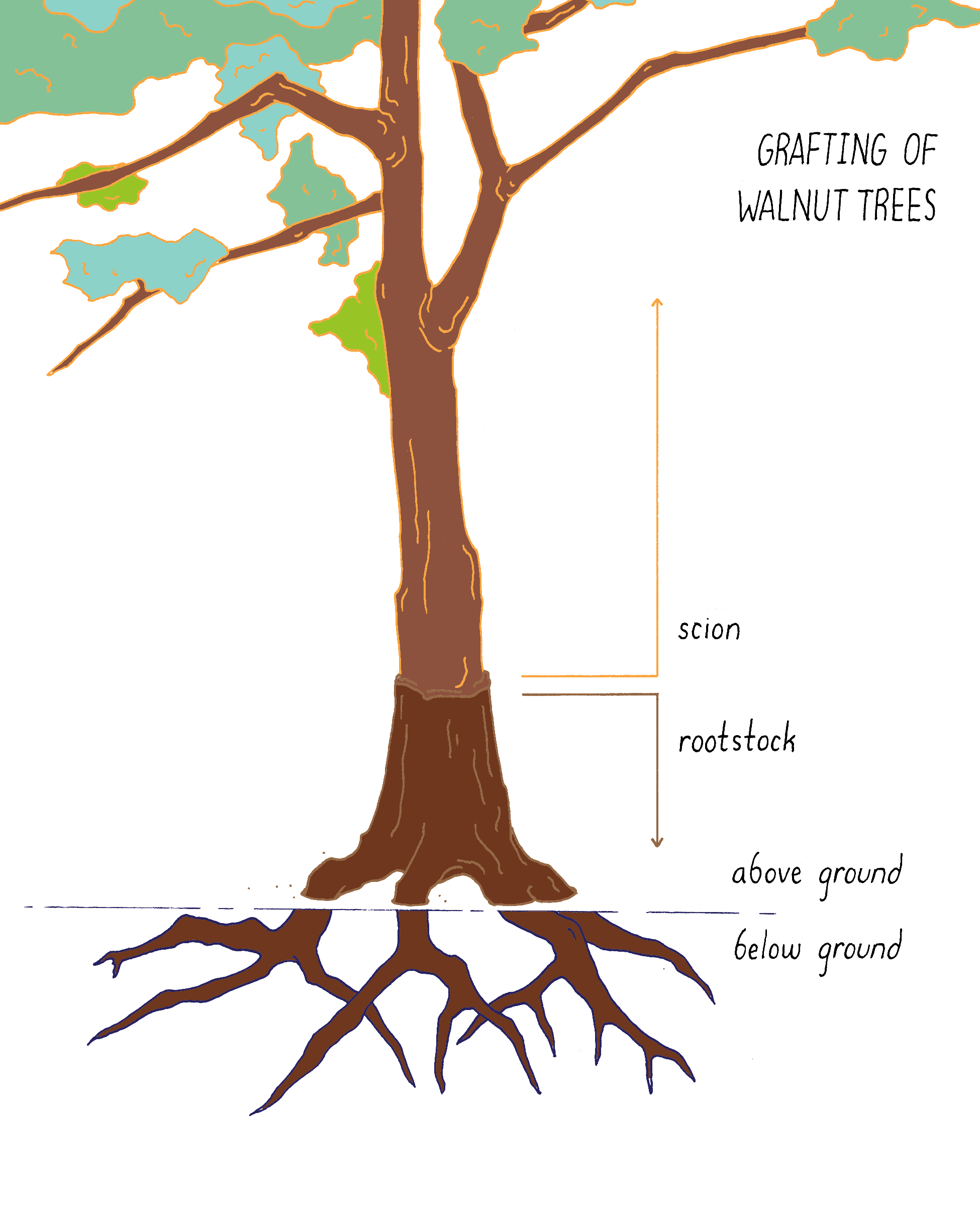
Induced Mutagenesis
In the early history of agriculture, farmer-scientists had to wait until spontaneous mutagenesis that is happening all of the time produced a desirable trait (and hopefully spot it). However, the frequency of spontaneous mutation is fairly low (see the Narrative on Mutagenesis by Koshland).
Now, scientists can accelerate this process by introducing higher mutation rates in plants. In this process, seeds are put into a mutagenic solution that induces random changes in the chemical letters of the DNA (see the Knowledge Overview in the Narrative on Mutagenesis by Koshland). Typically, about half of the mutagenized seeds will die since the mutations interfere with genes that are essential for plant life. The seeds from the surviving plants are collected, germinated, and surveyed for new traits by breeders. These induced-mutation techniques have been used in conventional breeding to generate ~2500 crops grown today. The outcome is the same as produced from naturally occurring spontaneous mutations, but the time scale for finding mutations that give rise to new phenotypes is dramatically reduced.
Modern plant genetics involving modern DNA methods
Marker-Assisted Breeding
Marker-assisted breeding (Figure 16) starts off with the same step – cross-pollination between two plants, each with a desirable trait(s). However, it differs in a more rigorous sorting of the offspring using tools of modern DNA analysis. In the previously described methods, the offspring from a cross-pollination are crudely selected based upon their phenotype, basically the “look” of the plant for a desirable trait. While it is easy to focus on the desired trait, sometimes other genes are introduced into that offspring that do not change the visual appearance but make it less robust or introduce other poor traits. How does one just get the “desired” gene for a particular trait but make sure that there is minimal excess baggage?
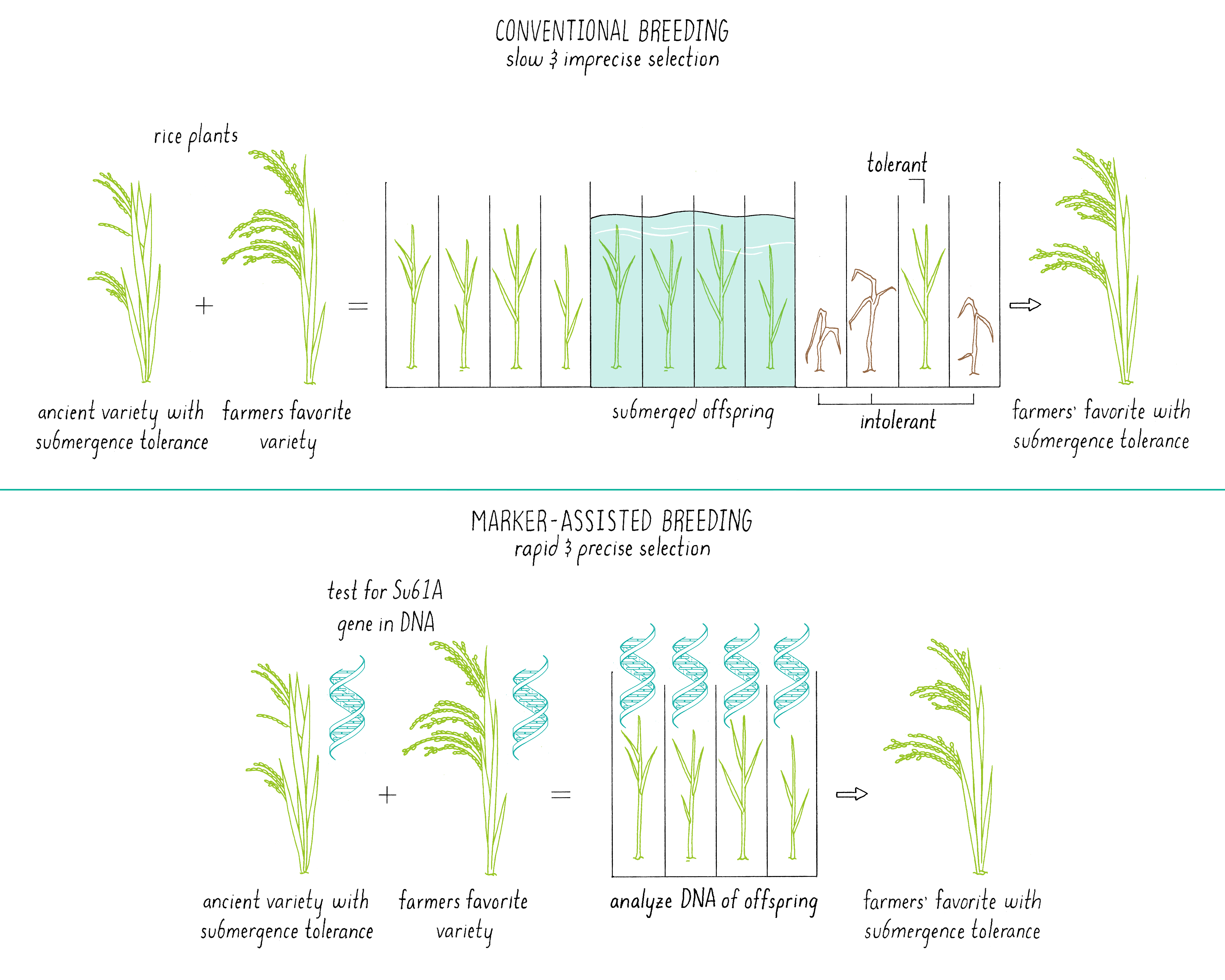
In marker-assisted breeding, the DNA of each offspring is analyzed (Figure 16). Only plants with the desired genetic composition are propagated further, saving time and labor. In addition, researchers do not need to grow each plant to maturity to find out what characteristics the plant will express. An inspection of the genome of the seed using selected DNA markers (see the Journey to Discovery) provides the needed information. In the case of the submergence tolerance gene (Sub1; see Journey to Discovery), it is much easier to determine if the offspring of cross between Sub1 rice and Swarna rice contained the Sub1 gene by DNA testing than by subjecting each plant to a 14-day submersion and a 7-day recovery period. The researchers could also select against traits that were derived from the ancient parent. Marker-assisted breeding is also discussed in Video 12.
Bioengineering
Today, scientists use modern genetic approaches to improve crops. Bioengineering is a process by which a gene, or a limited number of genes, can be introduced into a plant. In this case, the gene could be from the same plant species but also could be from another plant species, a bacterium or even an animal, as will be discussed in the Frontiers section.
Bioengineering in plants makes use of a soil bacterium called Agrobacterium tumefaciens (Figure 17). Normally, this bacterium is a plant pathogen that produces a “crown gall” which looks like a plant tumor. The bacterium infects a wounded root of a plant and then injects a plasmid that integrates some of its DNA into the host plant genome. The plant is then fooled into expressing genes for encoding enzymes that make metabolites that the bacteria needs for growth.
Scientists realized that they could use Agrobacterium tumefaciens as a means of delivering genes of interest from the bacterium to a plant. In this process, the disease-causing genes in the plasmid are removed. In place of these genes, scientists can introduce a new gene that they want to integrate into the plant genome. The bioengineered Agrobacterium tumefaciens can also be used to transfer DNA to plant cells in tissue culture, which then regenerate into a plant.
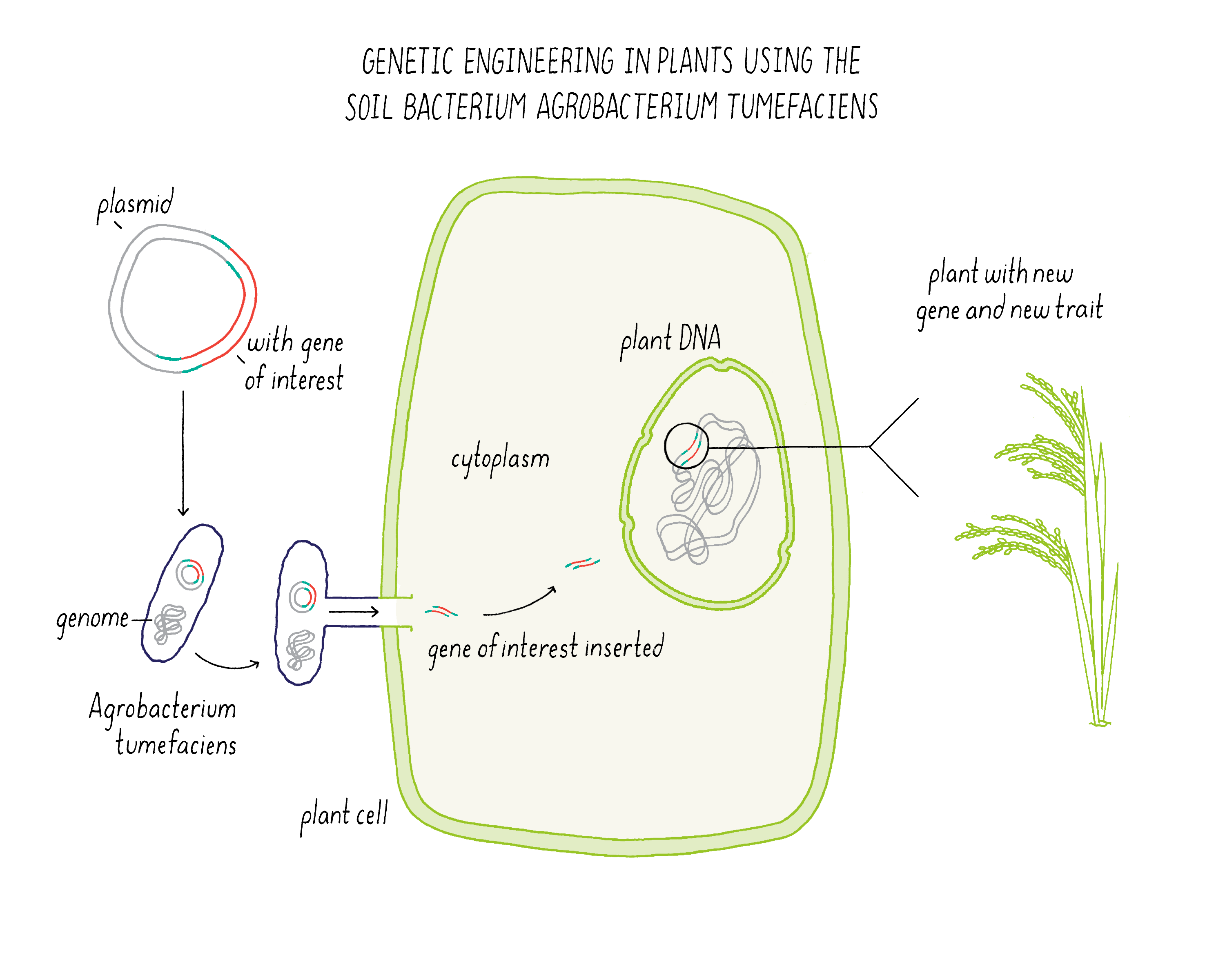
Explorer’s Question: In what ways does bioengineering differ from conventional breeding?
A. Conventional plant breeding mixes large sets of genes of unknown function, whereas bioengineering usually introduces only one or a few well-characterized genes at a time.
B. Conventional breeding allows gene transfer between closely related species. With bioengineering, genes from the same species or from any other species, even those from animals, can be introduced into a plant.
C. Bioengineering leads to plants that are sterile and cannot be replanted.
Answer: A and B are correct
Genome Editing
Unlike bioengineering, which integrates “foreign” DNA into plant cells, genome editing rewrites the DNA sequence in specific places of the plant genome. The method will not be covered here but is described in great depth in the Narrative on CRISPR by Barrangou. Genome editing can be used to introduce desirable mutations observed in wild species into elite varieties of crops, eliminating the need for years of breeding. Currently, researchers are trying to develop rice with improved traits using genome editing techniques.
The USDA has announced recently that it will not regulate some types of CRISPR-Cas9 genome editing, if only a few nucleotides are changed. It will likely regulate CRISPR-Cas9 mediated insertions of larger segments of DNA. The landscape of the use and regulation of genome editing will likely evolve over the coming years.
Summary of Plant Genetic Methods
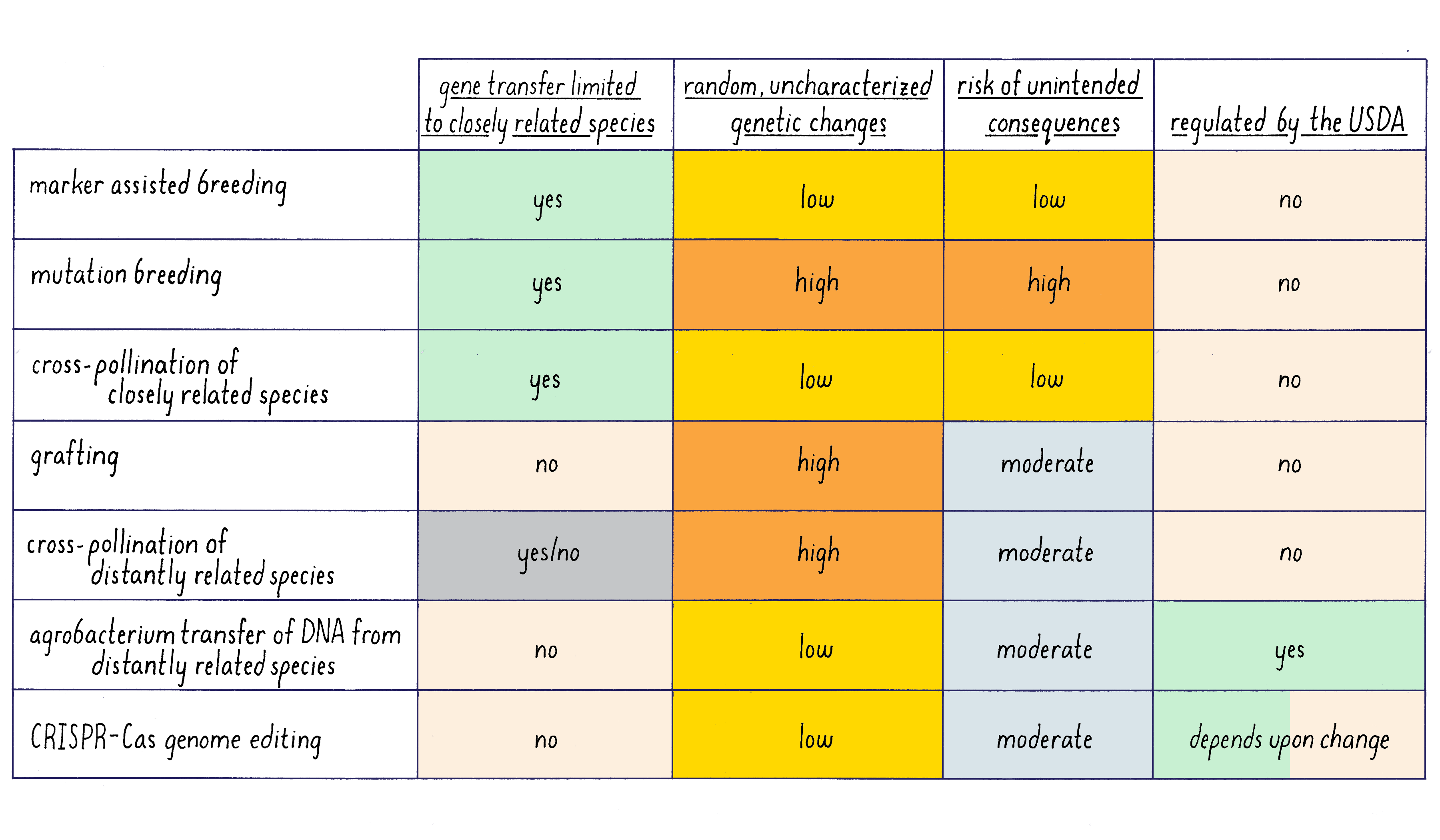
A summary of the feature of different methods of plant genetics is given in Figure 18.
Terminology associated with modern food
What is a genetically modified organism?
The term GMO has no precise scientific definition. As we have discussed in this section, virtually all foods that we eat have been genetically altered by humans using some form of genetic manipulation. A few exceptions include line-caught pacific salmon, chanterelle mushrooms gathered from oak woodlands, or wild blueberries from the forest.
Because there is abundance of confusion regarding the definition of “GMO” and because most foods do not contain entire organisms, the Food and Drug Administration of the United States does not use the term “GMO.” In 2018, the USDA announced that it would label some foods as “bioengineered” as discussed and defined below.
What is bioengineered food?
The term “bioengineered” (BE), signed into Law in 2016 by President Barack Obama, refers to crops that contain genetic material that has been modified through in vitro recombinant DNA techniques. For example, disease-resistant papaya that carries genes from a virus or golden rice (see the Frontier section) that carries genes from corn and bacteria are considered “bioengineered.” Under rules proposed in 2018, genome editing, if it results in minor changes of DNA sequences, will not be labeled as “bioengineered.”
After decades of careful study and rigorous peer review by thousands of independent scientists, every major scientific organization in the world, including the U.S. National Academies of Science and Medicine, has concluded that the bioengineered crops currently on the market are safe to eat and that the process of bioengineering is no more risky than older methods of genetic alteration (see References).
The Agriculture Marketing Service has proposed that the following BE foods already should be considered highly adopted. Their U.S. adoption rates according to 2016 ERS and ISAAA data are as follows:
Canola (90%), field corn (92%), cotton (93%), soybean (94%), and sugar beet (100%).
Commercially available BE foods – not highly adopted (< 85% in the United States) – are as follows:
Apple (non-browning cultivars), sweet corn, papaya, potato, and summer squash.
Explorer’s Question: Bt eggplant refers to a bioengineered eggplant that expresses a bacterial protein that acts as an insecticidal toxin. Are farmers that plant Bt eggplant and consumers that eat Bt eggplant exposed to more or to less chemical insecticides?
Answer: Less. Before the development of Bt eggplant, farmers sprayed their crops with a combination of chemical insecticides, many of which are known to be harmful to human health. Bt toxin comes from the bacteria Bacillus thuringiensis. Conventional and organic farmers have been applying the spores derived from B. thuringiensis as a liquid spray to a diverse array of crops since the 1920s. However, these Bt sprays are hard to find in Bangladesh, are expensive if available, and do not prevent the pest from getting inside the eggplant crop. With the introduction of Bt eggplant, Bangladeshi farmers have dramatically reduced the amount of chemical sprays on their crops.
What is a certified organic product?
Organic farming is governed by the U.S. Department of Agriculture (USDA) National Organic Program standards. Organic farming excludes the use of synthetic fertilizers and synthetic pesticides. Plants developed through hybridization, grafting, chemical or radiation mutagenesis, or treatment with the chemical colchicine can be certified organic by the NOP. Bioengineered crops are excluded from the NOP. As of 2018, the NOP had not yet ruled on whether or not crops developed through genome editing will be permitted.
Explorer’s Question: Does certified organic mean that the food is assessed for safety?
Answer: The NOP regulations do not address food safety or nutrition (https://www.ams.usda.gov/about-ams/programs-offices/national-organic-program).
Explorer’s Question: Does the NOP evaluate toxicity of the pesticides that organic farmers use?
Answer: The NOP relies on the EPA to assess toxicity of pesticides (the same agency that evaluates conventional pesticides). Organic farmers are allowed to spray pesticides that are harvested from the Earth such as copper sulfate or B. thuringiensis spores as described above.
Explorer’s Question: What regulations apply to the “Non-GMO” label?
Answer: Unlike the “BE” or “certified organic” labels, the “non-GMO” label is not regulated by the U.S. government. It can be purchased for a fee and applied to any product even water, salt, and sugar, which contain no DNA. The genetic testing process and the products sold with the “non-GMO” label are not publicly available, so it is not clear what exactly is being assessed in non-GMO labeled products.
Explorer’s Question: Do consumers pay more for foods with the “Non-GMO” or “GMO” label?
Answer: Yes. Consumers pay more for food labeled “Non-GMO.” The estimated price premiums paid by U.S. consumers for products labeled “Non-GMO” between 2009 and 2016 ranged from 10% to 62%” (https://www.sciencedirect.com/science/article/pii/S0306919218301131).
Explorer’s Question: Are “Non-GMO” foods promoting more harmful environmental practices?
Answer: The answer is complicated and requires a case-by-case analysis. For example, some “non-GMO” foods are derived from crops grown with older and more toxic chemicals. In an interview with NPR reporter Dan Charles, farmers noted that planting non-GMO beets would mean returning to older practices of spraying their crop every 10 days with a "witches brew" of five or six different weedkillers (https://www.npr.org/sections/thesalt/2016/05/12/477793556/as-big-candy-ditches-gmos-sugar-beet-farmers-hit-sour-patch).
Explorer’s Question: What are goals of sustainable agriculture?
A. Produce abundant, safe, and nutritious* food
B. Reduce harmful environmental inputs and minimize use of land/water
C. Protect the genetic makeup of native species
D. Enhance crop genetic diversity and foster soil fertility
E. Improve the lives of the poor
F. Maintain the economic viability of farming and rural communities
G. All of the above
Answer: G
Part III: Frontiers —
Golden Rice, a New Crop to Fight Vitamin A Deficiency
Humans require vitamins in their diet. A vitamin is an essential nutrient that we must acquire through food because we cannot make the compound in sufficient quantities.
Vitamin A is a chemical that is needed in growth and development and plays an important role in the immune system and vision. The consequences of vitamin A deficiency (VAD) are staggering. In 2012, the World Health Organization reported that about 250 million preschool children are affected by VAD and that providing those children with vitamin A could prevent about a third of all under-five deaths, which amounts to up to 2.7 million children each year. It is also the leading cause of preventable childhood blindness with a prevalence of 250,000–-500,000 of children becoming blind per year, according to resources in Wikipedia.
β-carotene (also called pro-vitamin A), which gives carrots their characteristic orange color, is converted to vitamin A in the liver (Figure 19). Rice does not contain β-carotene (pro-vitamin A), and thus, regions in the world where diets are based mainly on rice or other carbohydrate-rich, micronutrient-poor calorie sources, such as poor areas of Asia and Africa, experience high incidence of VAD. The dependence on rice by many people as the predominant food source is a primary problem leading to vitamin A deficiency.
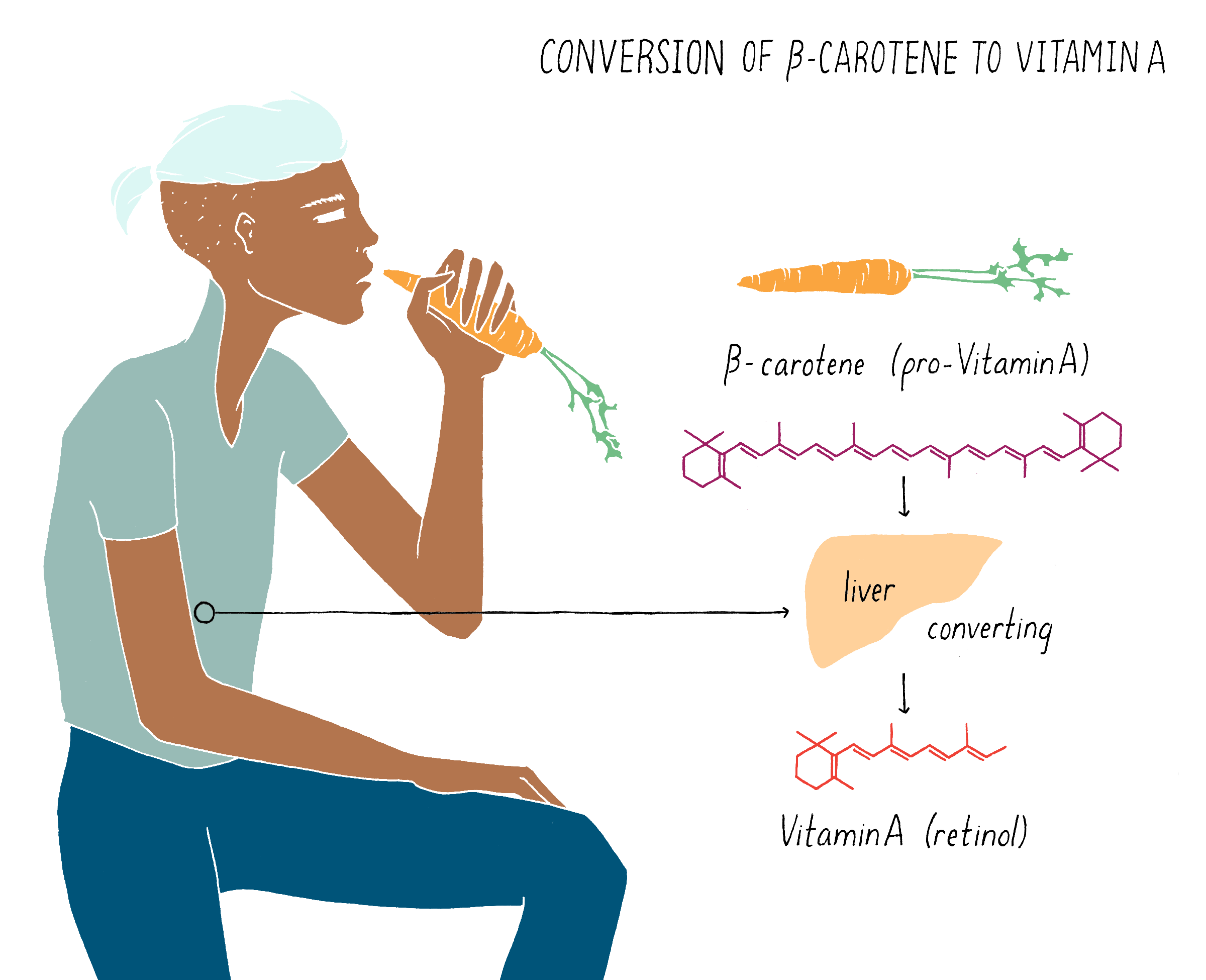
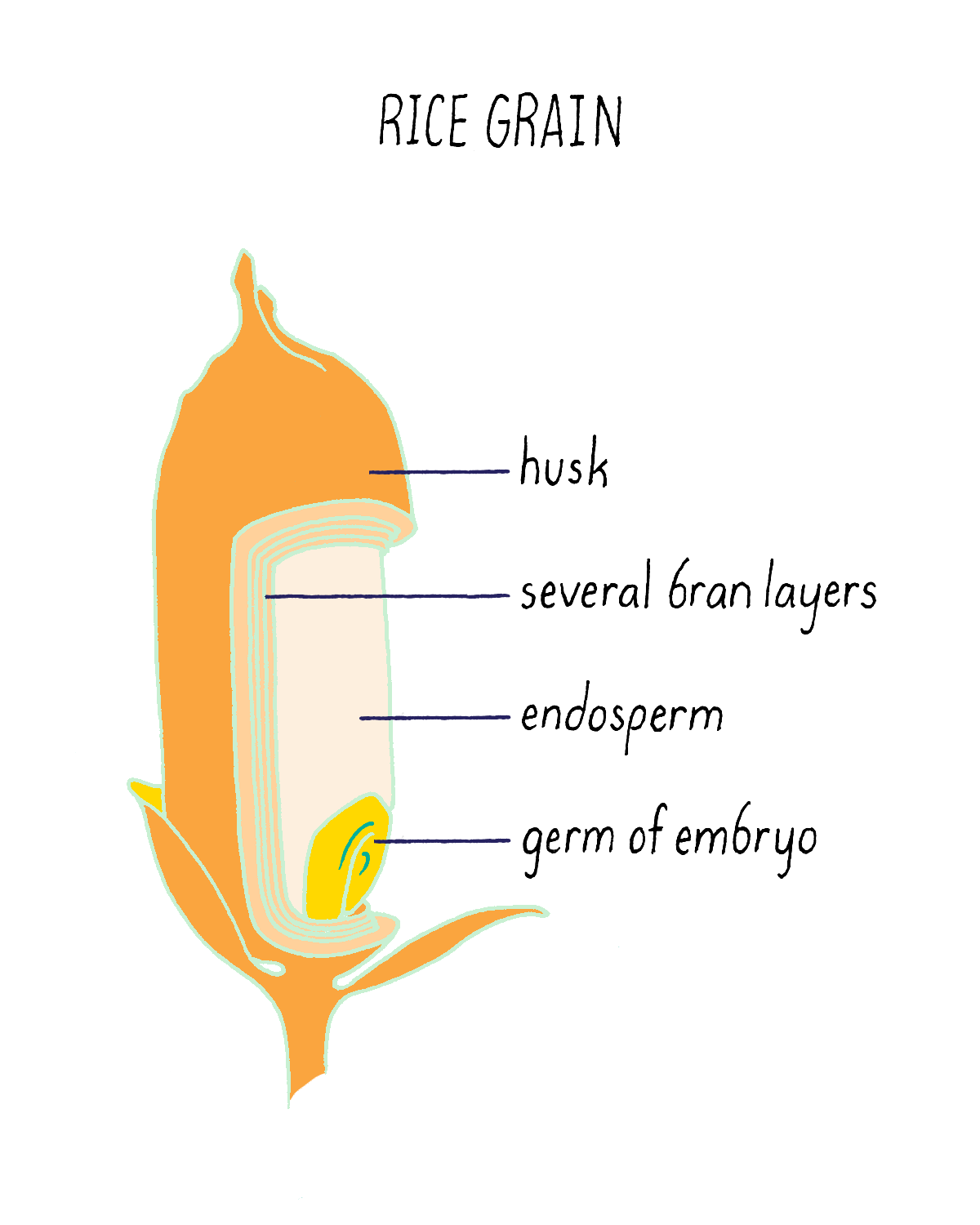
But what if rice could produce pro-vitamin A? Then, perhaps the global problem of vitamin A deficiency could be substantially reduced. The problem is not that the rice plant cannot make pro-vitamin A. It can, but only does so in the inedible leaves. The rice seed (see Figure 20), which contains an endosperm that acts as a food store for the developing plant embryo, is unfortunately devoid of pro-vitamin A. Thousands of existing rice varieties have been screened in the hope of finding one that might have pro-vitamin A-containing seeds. But no luck. Another strategy was needed.
Let’s start off with the question – how is pro-vitamin A synthesized and why is that rice seeds cannot make it? Pro-vitamin A is synthesized in series of chemical reactions called a “biosynthetic pathway.” Cells are busy factories, full of biosynthetic pathways that churn out numerous molecules that are essential for life. Nucleotides (building blocks of DNA), amino acids (building blocks of proteins), and phospholipids (building blocks of membranes) are all made by biosynthetic pathways, which are multistep reactions that start with simpler organic molecules. The series of production steps and the chemical intermediates involved in making pro-vitamin A are shown in Figure 21. Each reaction is catalyzed by a separate enzyme made by the cell.
Because scientists worked out all of the steps and enzymes required in the pro-vitamin A pathway, they could ask which ones were missing in the rice endosperm. What they discovered was that key enzymes in the middle of the pathway (enzymes 1–4 in Figure 21) are absent in the endosperm. The last two enzymes (5 and 6), however, are expressed in endosperm. Scientists then discovered that the genes for the first 4 enzymes are turned on in the leaf but turned off in the cells that give rise to the endosperm. In most cases, scientists do not have good strategies to switch on endogenous genes that are turned off. The workaround is to clone these genes and place them in front of a promoter that will always turn on these genes in the endosperm. Then, these promoter–gene complexes can be incorporated into the rice genome and the genes will be expressed in the endosperm of the rice seed.
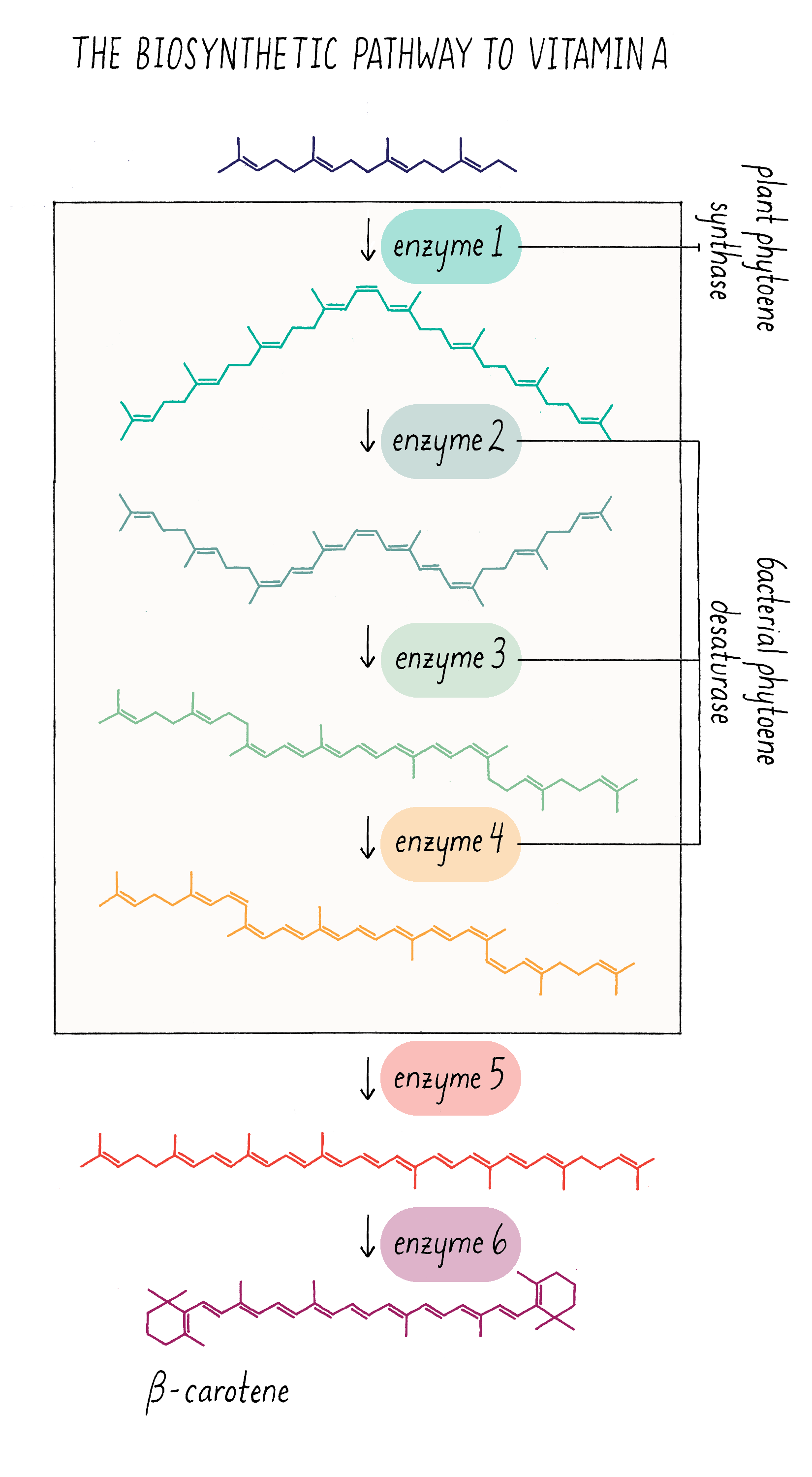
Cloning and delivering four genes by bioengineering is possible but not trivial. It would be easier to reduce the complexity, if possible. Fortunately, Peter Bramley made a discovery in the 1990s that a single enzyme from bacteria could carry out three of the four missing enzymatic steps! This enzyme (with the name phytoene desaturase) can catalyze the formation of the three double bonds and thus replace enzymes 2–4 in Figure 21. This suggested a two-step bioengineering strategy – introduce the gene for the first missing enzyme (enzyme 1, from daffodil or corn) and the gene for bacterial phytoene desaturase (equivalent to enzymes 2–4 from plants). For more information on bioengineering in plants, see the Knowledge Overview.
The first pro-vitamin A producing rice was reported in 2000. The resulting rice grains were gold colored and carotenoid rich, and hence, this new rice was dubbed “Golden Rice” (Figure 22). In 2005, a new bioengineered version was produced (Golden Rice 2), which contains 20 times more pro-vitamin A than its predecessor. Scientists at the International Rice Research Institute have introduced this trait into rice varieties grown by subsistence farmers. Studies showed that the pro-vitamin A in Golden Rice 2 was effectively converted into vitamin A in humans.

The first field trials in Bangladesh, where one in every five preschool children is vitamin A-deficient, were carried out in 2016, which yielded promising results. The researchers found that two months after harvest, the GR2 rice variety contained an average of over 10-μg pro-vitamin A per gram of rice. This amount is sufficient to meet 50% of vitamin A needs of people consuming rice in their daily diet. Studies have projected that widespread consumption of Golden Rice 2 would reduce vitamin A deficiency, saving thousands of lives. For example, a 2016 simulation indicated that if 70% of women and young children in Bangladesh, Indonesia, and the Philippines switched to golden rice, rates of VAD would be expected to drop from 93% percent to about 20% for women and 13% for children.
The positive effects of Golden Rice 2 are predicted to be most pronounced in the lowest income groups and come at a fraction of the cost of current supplementation programs. This low-tech, sustainable, publicly funded effort can complement other approaches, such as the promotion of home gardens with vitamin A-rich crops such as green beans and pumpkins, which are seasonally available.
Because Golden Rice is the product of bioengineering, some people view it with suspicion. Yet scientific studies indicate that the changes made in Golden rRce pose no risk to human health relative to non-bioengineered rice varieties currently available to consumers. Safety assessments are based upon scientific principles developed through expert international consultation over the last 20 years with agencies such as the World Health Organization (WHO), the Food and Agriculture Organization of the United Nations (FAO), and the Organization for Economic Co-operation and Development (OECD). In 2019, the Bangladeshi agricultural minister announced that the environment ministry will soon give clearance to Golden Roce for farm production. In the absence of unforeseen circumstances, Golden Rice will be made available to farmers and their families in the near future.
Closing Thoughts
With 11.2 billion people likely to be on Earth by 2100, the United Nations estimates that we will need to produce more food in the next 50 years than we have produced in the past 10,000 years (see References). And we need to do this while minimizing environmental impacts. We must either increase yields on our farms, change our eating habits, or reduce food waste, or better yet, do all of the above.
Because the amount of arable land is limited and what is left is being lost to urbanization, salinization, desertification, and environmental degradation, it is no longer possible to simply open up more undeveloped land for cultivation to meet production needs. Another challenge is that water systems are under severe strain in many parts of the world. The fresh water available per person has decreased fourfold in the past 60 years. Of the water that is available for use, 70% is already used for agriculture. Thus, increased food production must largely take place on the same land area while using less water.
Compounding the challenges facing agricultural production are the predicted effects of climate change (Video 13). As the sea level rises and glaciers melt, low-lying croplands will be submerged and river systems will experience shorter and more intense seasonal flows, as well as more flooding. Yields of our most important food, feed, and fiber crops could decline precipitously at temperatures much greater than 30°C, so heat and drought will increasingly limit crop production. In addition to these environmental stresses, losses to pests and diseases are also expected to increase. Much of the losses caused by these abiotic and biotic stresses, which already result in 30–60% annual yield reductions globally, occur after the plants are fully grown, a point at which land and water required to grow a crop already has been invested. For this reason, a reduction in losses to pests, pathogens, and environmental stresses is equivalent to creating more land and more water.
Thus, an important goal for genetic improvement of agricultural crops is to adapt our existing food crops to increasing temperatures, decreased water availability in some places and flooding in others, rising salinity, and changing pathogen and insect threats. Such improvements will require diverse approaches that will enhance the sustainability of our farms. These include more effective land and water use policies, integrated pest management approaches, reduction in harmful inputs, and the development of a new generation of agricultural crops tolerant of diverse stresses. These strategies must be evaluated in light of their environmental, economic, and social impacts – the three pillars of sustainable agriculture (Figure 23).
Genetic improvement of crops is a central component of sustainable agriculture. Genetic approaches that lead to the development of crops resilient to environmental stress are valuable to farmers, especially in the developing world. Because most of the world’s poorest people get their food and income by farming small plots of land, the availability of plant varieties with enhanced tolerance to disease pests and environmental stress can make a transformational difference in food security for these farmers.
With improved sequencing and engineering strategies, many new genes will be discovered and used to develop new crop varieties. A major goal of plant biologists is to survey the vast genetic diversity of plant species (much of which has been lost through years of conventional breeding) and to use this diversity to generate crops resilient to climate change that will help farmers support their families and communities.
Primitive plant domestication, bioengineering, and genome editing represent points on a continuum of new technologies developed through human endeavor and creativity. Which one of these tools is the most appropriate for agriculture? When the goal is a productive and ecologically based farming system, there are many interconnected possibilities. As the physicist and philosopher Jacob Bronowski pointed out 60 years ago,
“We live in a world which is penetrated through and through by science and which is both whole and real. We cannot turn it into a game by taking sides. … No one who has read a page by a good critic or a speculative scientist can ever again think that this barren choice of yes or no is all that the mind offers.” This idea extends to crop genetic improvement. Understanding both the specific application and the science behind it is important.
The hybridization methods, first described in 1899, the bioengineering of crops launched in 1996, and the genome editing of tomorrow are examples of a continuum of new technologies aimed at helping farmers produce food in a productive and ecologically based manner. The planting of genetically improved seed is a scale neutral technology, meaning that it is a technology that can be used by people with large farms and those on small farms.
Raoul, my husband who runs a small organic farm, and I believe that rather than focusing on how a seed variety was developed, we must ask what most enhances local food security and how can we provide safe, abundant, and nutritious food to consumers. We must ask if rural communities can thrive, if environmental damage can be reduced, if farmers can make a profit, and if consumers can afford the food. These are important issues. The solutions that we decide to implement must be driven by science as well as considerations of the well-being of humans and our environment.
Dig Deeper
Dig Deeper 1. Analyzing Sub1 genotypes
The only way to deduce the Sub1 genotypes of the F2 plants (shown in the right column in Figure 7) was to assess the submergence phenotypes of the next generation (the F3 progeny). To accomplish this, the F2 plants were self-pollinated, the "F3" seeds were collected, and plants were grown from these seeds for assessment of submergence tolerance. However, if we tested every F3 family from these F2 plants, the task would have been overwhelming. Since we did the tests in triplicates, we would have had to test the submergence phenotype of >120,000 plants (4000 F2 plants x 10 F3 progeny plants x 3 (triplicate) experiments). Furthermore, assessing submergence tolerance was time consuming and not easy (see Video DD1 on how submergence tolerance was tested). For this reason, Kenong first extracted DNA from each F2 plant and carried out DNA analysis to identify the plants that showed recombination events between markers A and D. In this way, he was able to reduce the number of F3 families that he needed to test for submergence tolerance from >4000 to 470. Testing submergence tolerance for F3 families from each of the 470 F2 plants was still a heroic effort! But using DNA markers as a first screening step, Kenong could save enormous time and effort and make the project manageable.
The F3 offspring were classified into three categories: (1) all F3 plants from an F2 self-pollinating parent were intolerant, (2) all F3 plants were tolerant, and (3) the F3 offspring segregated for both tolerance and intolerance. The full analysis is shown in Figure DD1. These results allowed us to infer the F2 chromosomal genotypes around the Sub1 region from the F3 phenotypes (shown in Figure 7). If all the F3 offspring were tolerant of flooding, this would mean that the corresponding parental F2 plant was homozygous in both chromosomal copies for the presence of the Sub1 gene. The recombinant chromosome must carry the Sub1 gene. If all the F3 offspring were intolerant, this would mean that the corresponding parental F2 plant was homozygous for the absence of the Sub1 gene. The recombinant chromosome lacked the Sub1 gene. If some of the F3 offspring were intolerant and some were tolerant, this segregating phenotype would mean that the F2 parent was heterozygous for the Sub1 gene (i.e., one chromosome of the F2 parent contained the Sub1 gene and the other chromosome did not). But which one, the recombinant chromosome or the nonrecombinant chromosome? In the F2 plants, if DNA makers on the non-recombinant chromosome were abcd, it came from the intolerant plant (all brown chromosomes) and lacked the Sub1 gene. So, the recombinant chromosome must have the Sub1 gene. On the other hand, if DNA markers on the non-recombinant chromosome were ABCD, it came from the tolerant plant (all green chromosomes) and contained the Sub1 gene. So, the recombinant chromosome must lack the Sub1 gene. Thus, by combining genotype analysis of the F2 plant with phenotype analysis of submergence tolerance of the F3, Kenong could deduce whether or not the recombinant chromosome possessed the Sub1 gene. Through such analysis of the recombinants shown in Figure DD1, he was able to pinpoint the location of the Sub1 gene to between markers B and C.
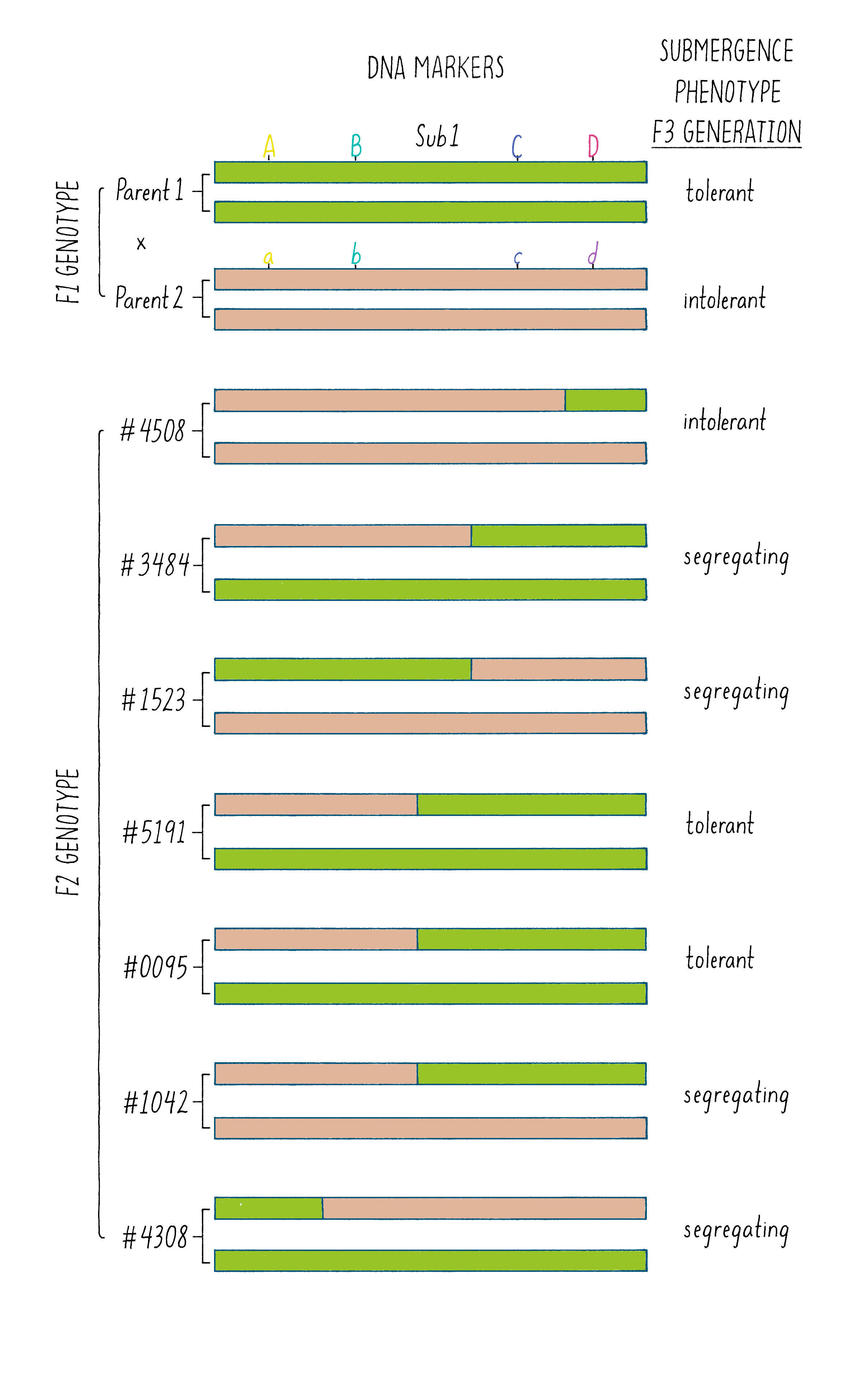
Based on this data, Kenong could calculate the frequency of recombination.
recombination frequency = (number of recombinant gametes / total number of gametes) × 100%
Only 5 recombinant progeny between markers B and C were found out of 4022 progeny; thus,
5 recombinant progeny/(4022 total number of progeny x 2 gametes per progeny) x 100(%) = 0.06%
(Recombination frequency can be expressed as either a decimal fraction (ranging between 0 and 1) or as a percent).
The recombination frequency of 0.06 indicates that the submergence tolerance gene is located in a 0.06 centimorgan (cM) interval on chromosome 9 between markers B and C. The 0.06 cM region contains 182,000 nucleotides (182 kb).
Dig Deeper 2. What is the mechanism of Sub1 action?
How does the single gene Sub1 allow the plant to survive underwater? Before Sub1 was isolated, researchers had hypothesized that submergence tolerance was related to the action of plant hormones. Hormones are molecules produced by plants and animals that regulate diverse biological processes. In plants, hormones regulate different aspects of plant growth. For example, the plant hormone gibberellic acid controls shoot elongation and the hormone ethylene is involved in ripening. Ethylene also accumulates in plants in response to environmental stress such as drought or submergence.
Researchers have observed that plants that are tolerant of submergence have reduced shoot elongation. Based on this observation, researchers hypothesized that Sub1 controlled the action of gibberellic acid. The sequence of Sub1 indicated that it was similar to proteins that serve as transcription factors, which are proteins that regulate a large set of genes (transcription factors are often called “master regulators"). When we compared Sub1 to other transcription factors, we found that it was most similar to transcription factors activated by the hormone ethylene.
In 2006, experiments led by Julia Bailey Serres and Takesh Fukao at UC Riverside tied these observations together. They showed that during submergence, elevated levels of Sub1A repress the activities of gibberellic acid. This repression in gibberellic acid results in a reduction in shoot elongation. Thus, during a flood, when the plants are submerged, the Sub1 plants do not try to grow out of the flood. In other words, the Sub1 plants adopt a “hold your breath" strategy; they stop growing until the flood subsides, conserving energy needed for survival. You can read the Bailey-Serres-Fukao paper describing this study at http://www.plantcell.org/content/18/8/2021.
Dig Deeper 3: Northern blot analysis of Sub1 gene expression
A gene is transcribed by RNA polymerase to make an mRNA, which is then translated by ribosomes into a functional protein. Therefore, by analyzing the levels of the mRNA, one can assess whether the gene is turned on and able to make a protein.
In this method (Figure DD3A), the RNA (including mRNA) is extracted from the rice plant. Then, it is pipetted into a well of an agarose gel and an electric current is applied (positive electrode on the bottom). The negatively charged RNAs then migrate through the gel from top to bottom and become separated by their size; smaller RNA can move through the agarose gel faster and end up closer to the bottom than larger RNAs. This is a way in which RNAs from the cell can be separated based upon the size. To better understand gel electrophoresis, see Video DD3. Also run your own simulated gels in the Gel Electrophoresis Activity, by Douglas.
Filter paper is applied to the agarose and the mRNA will be absorbed from the agarose to the filter paper in their same relative locations. The filter paper is then incubated with a radioactive DNA that is complementary in its sequence to a specific RNA, in this case the Sub1A mRNA. Because of this complementary, the radioactive DNA will hybridize (form a DNA-RNA double helix) with the Sub1A RNA in its migration position on the filter paper. Then, the filter paper is washed off of unhybridized radioactive DNA, and the washed filter paper is exposed to an X-ray filter. The radioactivity from the DNA-RNA hybrid will appear as a dark band on the X-film when it is developed. From the position of the band, one can assess the size of the RNA and from its intensity one can infer the amount of RNA that hybridized with the DNA.
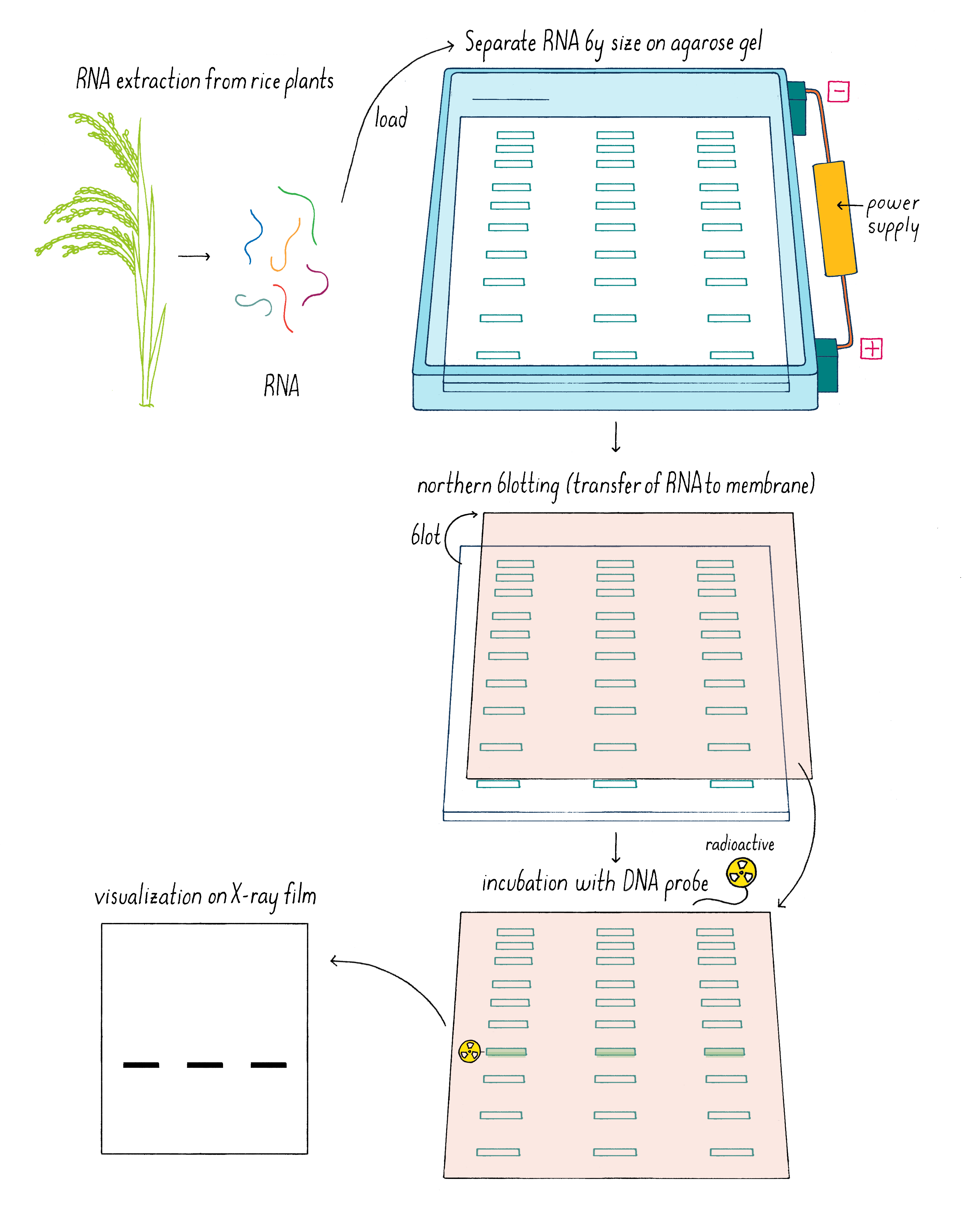
Now let’s see what the actual Northern blot looked like for Sub1A (Figure DD3B). In this case, RNA was extracted from different rice plants after they were submerged. “0" time means the RNA was extracted just before flooding. The data show that the Northern blot band gets more intense after one hour of submergence and increases over the next 4 hours, meaning that the rice plant is making more Sub1A mRNAs. This photograph is cropped to feature just the region of the Northern blot with Sub1A mRNA (the whole blot shows no other bands, indicating that the radioactive DNA probe was highly selective for Sub1A mRNA and did not react with other mRNAs).

Where did the strange name “Northern blot” come from? Before the Northern blot, a scientist named Ed Southern developed the first technique of transferring DNA (instead of RNA) from an agarose gel to filter paper and hybridizing with a radioactive DNA probe. It got named “Southern blot” after the discoverer. Two years later, J.C. Alwine developed the analogous method for RNA as described above. Instead of calling it the “Alwine blot,” he named it in the modest and more amusing fashion as a “Northern blot.” The trend continued. Later a method of transferring protein to paper and probing with an antibody was developed, and it was called a “Western blot.” The latest “Eastern blot” probes for post-translational modifications of proteins.
References and Resources
Guided Paper
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., Ismail, A.M, Bailey-Serres, J., Ronald, P.C., and D. J. Mackill. (2007). Sub1A is an ethylene-responsive-factor-like gene that confers submergence tolerance to rice. Nature, 442: 705–708.
This paper describes the discovery of a gene that controls the ability of plants to resist being submerged. As a result, new strains of rice can be produced that are protected against otherwise damaging floods, increasing food security for farmers.
DownloadThis is the paper featured in the Journey to Discovery.
References
- Dubock, A. (2016). Biofortified golden rice as additional intervention for vitamin A deficiency (VAD). In Saskai, T (ed.) Achieving sustainable cultivation of rice Volume 1. Burleigh Dodds Science publishing, Cambridge, UK.
Additional reading on golden rice featured in the Frontiers section.
- Fukao, T., Xu, K., Ronald, P.C., and Bailey-Serres, J. (2006). A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell, 18(8): 2021–34.
The paper outlining the study is described in Dig Deeper 1.
- Reports on the labeling and safety of Bioengineered Foods
- National Academies of Sciences, Engineering, and Medicine, Genetically Engineered Crops: Experiences and Prospects. Washington, DC: The National Academies Press, 2016, 420 p.; Statement by the AAAS Board of Directors on Labeling of Genetically Modified Foods. Washington, DC: American Association for the Advancement of Science, 2012. Available from: https://www.aaas.org/sites/default/files/AAAS_GM_statement.pdf
- Institute of Medicine, Committee on Identifying and Assessing Unintended Effects of Genetically Engineered Foods on Human Health, Safety of Genetically Engineered Foods: Approaches to Assessing Unintended Health Effects. Washington, DC: The National Academies Press, 2004, xvii, 235.
- Information on human population growth and climate change http://www.peopleandtheplanet.com/
Resources
- How Does Rice Grow: https://www.youtube.com/watch?v=kxAEiHCErSA
This short video explains the cultivation of rice and how it is harvested.
- Time-lapse of Sub1 Rice Growing in a Flooded Field: https://www.youtube.com/watch?v=DJsNwYX1Nc0
In this short time-lapse video, rice growers demonstrate how rice varieties that carry the Sub1 genetic locus can survive a flood compared with a variety lacking the Sub1 genetic locus.
- The Future of Rice Farming & the Importance of Partnerships in Rice Research: https://www.youtube.com/watch?v=FfjW_j9Wp_M
This video features Robert Zeigler, IRRI director general, and Achim Dobermann, IRRI deputy director for research, discussing how collaboration between researchers has improved rice crops as well as how new technologies such as CRISPR can push the research farther in the future.
- Genetic engineering report: https://nas-sites.org/ge-crops/2016/05/16/report-in-brief/
This National Academy of Sciences report presents a summary of knowledge of genetically engineered crops.
- Golden Rice: http://www.goldenrice.org/
This webpage provides a wealth of information regarding the development, cultivation, and future directions of golden rice.
- Homologous Recombination Videos: https://www.youtube.com/watch?v=yhZlfTr-ZEs and https://www.youtube.com/watch?v=5638I3fGuW4
These two short videos/animations discuss and provide visuals for the process of homologous recombination.
Activity
Learn the principles of agarose gel electrophoresis yourself and run your own simulated gels in the Gel Electrophoresis Activity, by Douglas.