CRISPR-Cas
From Bacterial Adaptive Immunity to a Genome Editing Revolution
Rodolphe Barrangou
 Rodolphe Barrangou
Rodolphe Barrangou
Dr. Barrangou spent 9 years in R&D at Danisco and DuPont in the Food Industry, and is the T. R. Klaenhammer Distinguished Professor in Probiotics Research in the Department of Food, Bioprocessing and Nutrition Sciences at North Carolina State University. He teaches an introductory undergraduate course on bioprocessing (the use of biological processes and materials to manufacture products) and biopharmaceutical sciences. His lab focuses on the functions of CRISPR-Cas systems, and their applications in bacteria used in food manufacturing. Rodolphe has received several awards for this work on CRISPR, including the Canada Gairdner Award, and has been involved in several start-up companies using CRISPR-based technologies.
Summary
Humans are plagued by viruses, which cause illnesses from the common cold to AIDS. However, humans are not unique in their need to fight viruses. Even bacteria are preyed upon by viruses called bacteriophages, which are the most ubiquitous and abundant biological entity on our planet. Viruses and bacteria are engaged in an evolutionary arms-race for survival, which has given rise to a diverse set of defense systems that enable bacteria to evade phage predation. Similar to adaptive immunity in humans, bacteria can capture and store a molecular memory of a prior encounter with a virus in their genomes, and this vaccination" allows bacteria to seek and destroy the nucleic acids of subsequent related invaders. The discovery of this bacterial immune system, called CRISPR-Cas, emerged unexpectedly from putting together clues about mysterious DNA sequences in bacterial genomes, and from the need of the food industry to keep bacterial yoghurt and cheese cultures free of viral infection. After this discovery, scientists learned how to repurpose the molecular machinery from these immune systems to edit the sequence of genomes, including those of humans. In addition to creating a new research technology that is sweeping the globe, the ability to rewrite genomes has monumental implications for medicine (human gene therapy), agriculture (next-generation plants and livestock), biotechnology (engineering of microbes for the synthesis of bioproducts), and beyond.
Learning Overview
Big Concepts
The CRISPR-Cas bacterial immune system allows bacteria to selectively remember the nucleic acid sequences of prior viral invaders and defend themselves against future viral attacks. Scientists have exploited knowledge of the molecular machines involved in the bacterial immune system to develop one of the most powerful tools ever created for biotechnology—the ability to edit the genome of any organism.
Bio-Dictionary Terms Used
B Cell, Bacterial Colony, Bacterial Strain, coding and non-coding, DNA, Enzyme, Fermentation, Gene, Genome, Haploid/Diploid, Homologous DNA Sequence, Human Immunodeficiency Virus (HIV), Immunity, Locus, Microbiome, Pathogen, Bacteriophage (phage), Plasmid, Prokaryote (bacteria), Protein, Reading Frame, DNA Replication, RNA, RNA interference, T Cell, Transcription, Virus, Vaccine
Terms and Concepts Explained
Bioinformatics, CRISPR, Cas Genes, Cas9, Duchenne Muscular Dystrophy, Genome Editing, Gene Knockout, Single-Guide RNA, non-homologous end joining (NHEJ), homology-directed repair (HDR)
Introduction
-
Bacteria are invaded and destroyed by bacteriophage (phage) and have evolved mechanisms to protect themselves.
-
This Narrative describes the discovery, mechanism and applications of one such bacterial defense system, CRISPR-Cas.
Part I: Journey to Discovery – CRISPR-Cas Systems Provide Adaptive Immunity In Bacteria
-
Microbes have been used to produce food products through fermentation for millennia.
-
This Journey to Discovery was stimulated by a very pragmatic problem faced by a food company called Danisco, Inc. How can bacterial starter cultures for yoghurt be protected against viruses (bacteriophage)? And why did some bacteria appear to be resistant to viral attack?
-
Scientists discovered a peculiar region in the bacterial genome with a nucleotide sequence that was repeated over and over again; they later called it the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), but its function remained mysterious for decades.
-
An intriguing gene called Cas9 was found adjacent to CRISPR. Bioinformatics suggested a possible function of the gene in the interference of nucleic acid function.
-
In between the CRISPR repeats were variable nucleotide sequences called “spacers.” Originally thought to be random and uninteresting, scientists discovered that the spacer sequences were similar or identical to sequences found in bacteriophages.
-
Scientists at Danisco, Inc. had access to a large amount of sequence data of different yoghurt culture strains of bacteria and the phage to which they were exposed. They found that the CRISPR spacer sequences in the bacteria were similar or identical to the sequences of phage that attacked them. This finding suggested that perhaps CRISPR plays a role in acquiring immunity to attack by a particular phage.
-
Scientists now performed experiments to test the hypothesis that CRISPR plays a role in phage immunity. They challenged bacteria with a particular phage strain. Survivors of the attack had acquired new CRISPR spacer and those spacers matched the sequence of the phage that attacked them.
-
They then intentionally manipulated the spacer region by genetic engineering with the following outcomes: 1) Adding a spacer, phage resistance was acquired; 2) removing a spacer, resistance was lost; 3) swapping spacers between two strains, their respective phage resistance was switched too.
-
The scientists also tested the function of the Cas9 gene. If the gene was “knocked out,” phage resistance was lost.
-
These experiments established the biological function of CRISPR-Cas systems as providing sequence-specific adaptive immunity against viruses in bacteria.
-
Shortly thereafter, several groups showed that the CRISPR locus produces RNAs and that Cas9 is a nuclease that, under the instructions of the CRISPR RNAs, targets and cleaves the bacteriophage DNA, destroying its replication potential.
Part II: Knowledge Overview – CRISPR-Cas Systems
-
CRISPR-Cas9 provides an immune system for bacteria. The memory of a prior phage attack is maintained by storing a small segment of the phage DNA sequence in the bacterial genome (at the CRISPR locus). The Cas9 enzyme uses this information to cleave the DNA of the same phage if it tries to invade again.
-
There are three main steps in the bacterial immune response. In the first step (acquisition), a segment of the invasive DNA is copied (creating a “spacer”) and then inserted along with a new CRISPR repeat at the end of the CRISPR array.
-
In the second step (expression), the CRISPR array is transcribed into a long precursor CRISPR RNA molecule, which is then cleaved into small CRISPR RNAs (crRNAs).
-
In the third step (interference), a crRNA and a second RNA (tracrRNA) form a complex with the protein Cas9. The Cas9-RNA complex scans along DNA randomly; if it finds a sequence that matches the crRNA, it stops and then cleaves the target DNA.
-
The Cas9 enzyme has been turned into a programmable genome editing tool. Also see the Key Experiment on CRISPR-Cas9 by Doudna.
-
Cells will repair a double-strand break in DNA using two types of DNA repair: non-homologous end joining (NHEJ) and homology-directed repair (HDR).
-
CRISPR-Cas9 can be programmed to create a double-strand break at a specific sequence in the genome. Scientists can also introduce a “repair template” that the repair machinery can use to repair the break. The repair template can be designed in such a way that a new sequence (an “edit”) is introduced into the genome during repair.
-
CRISPR-Cas9 editing has many applications. It can be used to change one (or a few) nucleotide(s) to another (e.g., to correct a disease mutation), insert a new piece of DNA, or delete a piece of DNA.
-
The steps in the genome-editing experiment are described.
Part III: Frontiers – Uses Of CRISPR-Based Technologies
-
CRISPR-based technologies have democratized genome editing and can be easily implemented in animal, plant, and microbial cells across the tree of life with implications in medicine, agriculture, and biotechnology.
-
Duchenne Muscular Dystrophy (DMD), a severe disease of muscle degeneration, is a recessive genetic disorder arising from a mutation in a gene on the X chromosome. The disease occurs primarily in boys due to their single copy of the X chromosome.
-
Dogs with a similar DMD mutation are being used as a model to study and treat the disease.
-
A study is presented in which a virus is used to deliver Cas9, a sgRNA, and a DNA repair template to the muscle of dogs with this disease.
-
Results show that the defective DMD gene in many, but not all, of the muscle cells is corrected by CRISPR-Cas9 and a healthy appearance of the muscle is restored.
-
Examples of how CRISPR-Cas9 genome editing is being used in agriculture and livestock are discussed.
Closing Thoughts
-
CRISPR is a great example of how powerful new technologies emerge from asking basic questions about how life works, in this case, starting with observations of mysterious sequences in the genome of bacteria.
-
The ability to edit genomes with ease through CRISPR-Cas technologies raises many important ethical questions on how this technology should be used in humans and other species.
Guided Papers
Barrangou, R., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709-1712.
This paper describes how the CRISPR-Cas system functions as an adaptive bacterial immune system. This knowledge allowed subsequent development of CRISPR-Cas as a versatile genome editing method.
DownloadAmoasli, L. et al (2018). Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362: 86-91.
The authors use CRISPR-Cas gene editing to correct the effects of mutations in the gene encoding dystrophin that cause Duchenne muscular dystrophy (DMD). This paper shows the efficacy and safety of the CRISPR-Cas system in large mammals.
DownloadIntroduction
As humans, we are mindful of the many viruses that surround us. Our history is plagued with catastrophic viruses, and recent viral outbreaks are a reminder of their impact on human health, including disabling or life-threatening diseases (e.g.,HIV and flu), persistent infection (e.g., herpes and papillomavirus), food poisoning (e.g., norovirus), and animal disease (e.g., H1N1). All living organisms are subject to predation by viruses, which are the most numerous biological entities on the planet. Indeed, viruses can be detected in all environmental conditions and niches, sometimes in astonishingly high numbers: a sip of ocean water contains more viruses than the number of humans on Earth. Bacteria suffer the greatest sheer number of viral attacks due to their large numbers. The viruses that attack bacteria are called bacteriophage (or phage).
Like other organisms, viruses have a genome consisting of nucleic acid (DNA or RNA), which is surrounded by a protein shell (the capsid) and sometimes by a membrane. However, the difference between viruses and other living organisms is that viruses neither can replicate on their own nor do they carry out metabolic activities. Rather, they must invade a host cell and subvert some of the host machinery for the replication and manufacturing of more viruses (Figure 1). A single infection typically yields dozens of new viruses from a single host. The compact machinery and rapid replication of viruses makes them robust, efficient, and fast-evolving molecular machines. Bacteriophages are also deadly to their hosts: to release their new copies, phage usually lyse and ultimately kill the bacterium.

As you might imagine from their predator–prey relationship, viruses and their hosts are engaged in an evolutionary arms race for survival. While viruses try to gain command of the host machinery, hosts develop an arsenal of defense and immune systems that enable them to overcome viral predation. Indeed, viruses need to evolve quickly to adapt to their host, while the host needs to mount an adaptive response to prevent viral predation or escape viral infection. Given the pace at which bacteria grow and replicate (a single bacterium can become a billion overnight), the bacteria-phage arms race is unraveling at the most frenetic pace and scale of all predator–prey relationships on earth.
In humans, the adaptive immune response is a powerful way of fighting viral (or bacterial) infection. For example, a flu vaccine delivers an inactive virus and some of your white blood cells retain a memory of this viral signature. Consequently, these cells are ready to be quickly mobilized in case the real (or a closely related) virus shows up later. (It is worth noting that mutations rapidly change the DNA sequence of the flu virus, which is why flu vaccines are only partially effective.) For many years, scientists considered bacteria to be simple and incapable of mounting a specific immune response to viruses. However, we have had a long track record of underestimating and under-appreciating the sophistication of bacteria (see also the Narrative on Quorum Sensing by Bassler). This Narrative is about a bacterial adaptive immune system called CRISPR-Cas.
In the Journey to Discovery, I will trace the key discoveries that led to our understanding of CRISPR-Cas as a bacterial immune system. The journey, like so many in science, involved several investigators contributing various clues and pieces of the puzzle. CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats, which is a mouthful for describing a mysterious set of repeated nucleotide sequences. Although these repeating sequences showed up in the genomes of many prokaryotes, their function remained elusive for nearly two decades after they were initially observed. Eventually, a series of clues led several inquisitive groups to posit that they may constitute a part of an immune system against viruses. This hypothesis naturally led to experiments that my group conducted, demonstrating that CRISPR-Cas provides adaptive immunity in bacteria.
In the Knowledge Overview, I will explain how several types of CRISPR-based immune systems in bacteria work. When bacteria encounter a viral attack, they acquire a piece of viral DNA to vaccinate them against future attacks by similar viruses. These systems can then thwart subsequent attacks by selectively cleaving invasive nucleic acid sequences that match the piece of DNA acquired during the vaccination process. I will discuss how various CRISPR-Cas systems generate different types of damage in DNA and RNA. I will also explain how these machines can be reprogrammed and transplanted into other organisms for flexible targeting and cleavage of DNA, and how the resulting DNA breaks can be repaired in ways that enable the rewriting of the DNA sequence in a genome (known as genome editing). I also encourage you to read the Key Experiment by Doudna, which describes a critical result that opened up the possibility of creating a programmable CRISPR-Cas system for editing any sequence in the genome. In the Frontiers section, I will discuss how CRISPR technologies are being exploited by thousands of scientists to alter the genomes of animals, plants, and microbes and will pick a couple of examples to highlight this exciting new technology and its impact on medicine, agriculture and biotechnology.
From what started as curiosity about strange sequences in bacteria, CRISPR has revolutionized biology in the past decade and has now become a household word. The implications of CRISPR-fueled genome editing on medicine, agriculture, and biotechnology are staggering. With this powerful new technology comes profound implications for society, encompassing ethical concerns, regulatory processes, and industrial challenges, which I will return to in my Closing Thoughts. In addition to shedding light on how we came upon this disruptive discovery and how it is impacting many fields of science, this Narrative also reveals how science relies on the contributions of many individuals and how a scientific journey proceeds with a blend of eureka moments and mistakes, all critical to the scientific process itself.
Part I: The Journey to Discovery —
CRISPR-Cas systems provide adaptive immunity in bacteria
The Problem
Eating is perhaps the most basic human need—we have domesticated the land and oceans, cultivating many plant and animal species throughout history to feed ourselves (also see Narrative on Plant Genetics by Ronald). Despite all of our scientific knowledge and technological advances, we still struggle to feed humanity a plentiful, safe, tasty and health-promoting diet. Although most people are very familiar with food as a product and a commodity that we consume several times a day (hopefully), consumers typically have a very limited understanding of where their food comes from, let alone how it is made or scientifically formulated. Indeed, humankind used fermentation for millennia to preserve and process foods without any understanding of the microbiological organisms and processes that drive it. Microbes actively assist in the preservation and genesis of enjoyable products, including converting grapes into wine, apples into cider, barley into beer, milk into cheese, cabbage into sauerkraut, or cucumbers into pickles. For more than a century, Louis Pasteur and subsequent food microbiologists have been working on harnessing microbes for the genesis of tasty foods. However, occasionally, fermenting organisms also get infected by viruses with sometimes dire consequences.
In 2005, I was an industrial food scientist at Danisco, Inc., and we had a problem that was costing our company time and money. Yoghurt and cheese are made through fermentation of milk by starter cultures of bacteria and maintaining healthy cultures is the heart of a successful business (Figure 2).

However, many of our yoghurt cultures were occasionally dying, victims of bacteriophage that can infect and kill the bacteria. However, some of the bacteria cultures survived. Why was it that some bacteria survived while others succumbed to the virus? The answer to this question could have very practical consequences for our company, which provided starter cultures to the dairy industry, and our customers, large yoghurt and cheese industrial manufacturers. Rather than addressing a specific fundamental question or hypothesis, the practicality of the issue at hand provided a sense of urgency and a real industrial problem to solve—how do some bacteria survive phage attack?
In this Journey to Discovery, our story begins with a puzzling observation of strange sequences in the DNA of bacteria, which were completely mysterious at the beginning and of interest to very few people. But then a trail of clues began to emerge from several scientists that ultimately led to the discovery of the natural function of CRISPR as an "immune system" that protects bacteria against viruses. This provided an answer to the problem that our company faced of understanding how certain bacterial cultures survived phage attack. Eventually, those studies laid the foundation for today’s "CRISPR craze," the most disruptive new biotechnology to emerge in the past two decades.
Clues
Clue 1: CRISPR sequences in bacterial genomes
The first clue in the CRISPR story can be traced back to 1987. At that time, Ishino and colleagues in Japan serendipitously stumbled upon an unusual DNA sequence composed of five identical sequences of 29 nucleotides (Figure 3). They noted the sequence in one of their publications but said that it had no known "sequence homology … and biological significance." This observation received little attention and was ignored and overlooked for many years.
The curious ~29 nucleotide repeats resurfaced a decade later in the late 1990s when the genomes of many prokaryotes were being sequenced. Only a few people, however, were curious enough about them to point them out. Francisco Mojica was particularly noteworthy among those individuals, since he not only noticed them, but also chose to pursue them for his research. Mojica found these sequences in more than 20 different types of bacteria. Since they were so prevalent, they must be doing something important, but what?
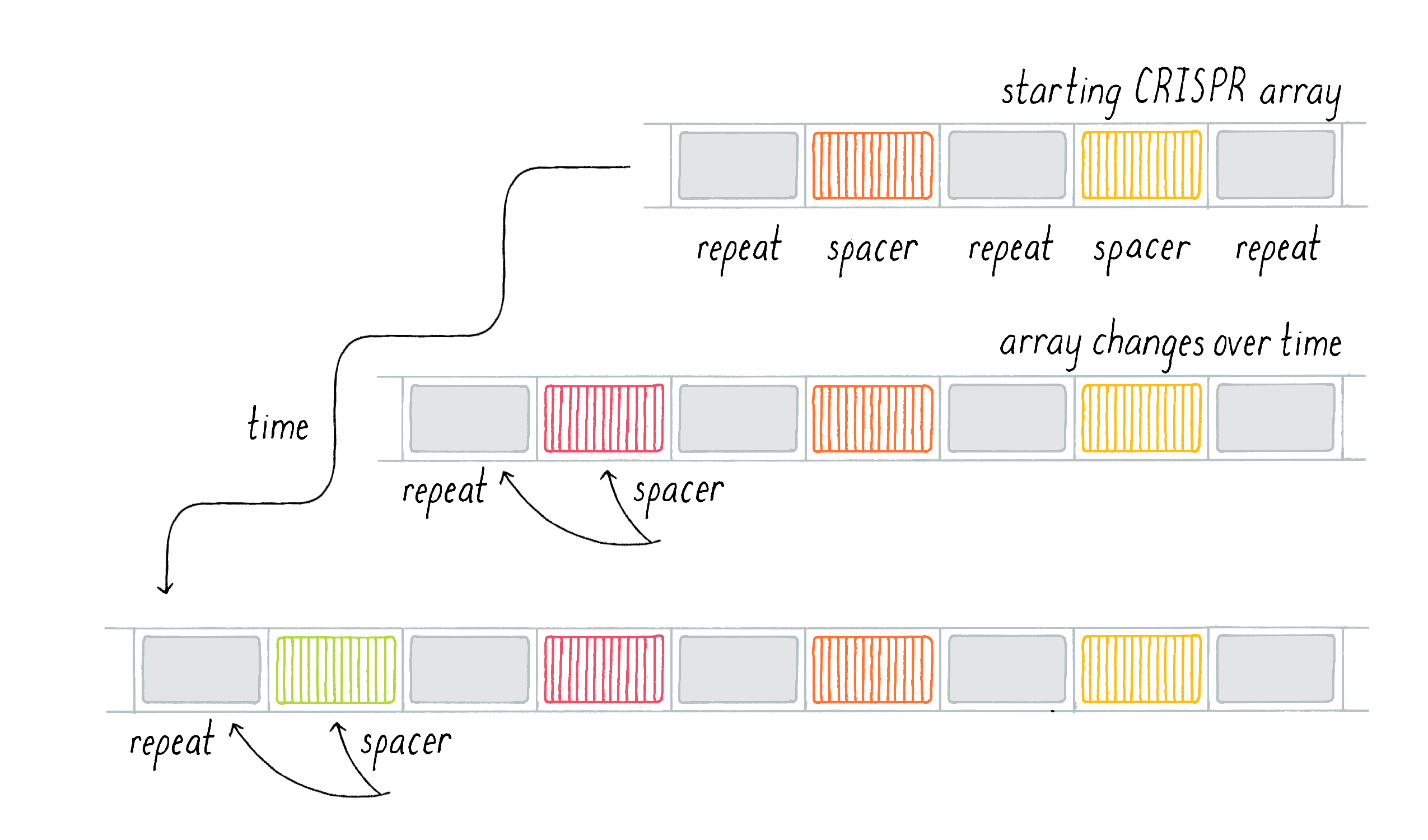
Researchers needed a unifying name to refer to these mysterious sequences. In 2002, the field converged upon the acronym CRISPR for Clustered Regularly Interspaced Short Palindromic Repeats. CRISPR encompasses short stretches of repeated DNA that are clustered together at a particular genetic locus and are partially palindromic: they read the same read backwards and forwards, as with the words "mom" or "racecar." These intriguing DNA repeats are typically separated by seemingly random, at least at that time, DNA sequences named "spacers" (Figure 3, CRISPR sequences). The name "Spacers" make these sequences sound boring, but they will emerge as important players later on (see Clue 3).

Clue 2: CRISPR sequences are paired with Cas genes
Genomes are composed of "coding" and "non-coding" regions. The coding regions are elements of genes and are transcribed into RNAs, which in turn are translated into proteins (see Central Dogma). Some of the non-coding regions in the genome are transcribed into RNAs that do not make proteins but serve functional roles in the cell. We will see later that the CRISPR sequences make such non-coding RNAs.
The genes are usually easier to spot in the enormously long sequence of bases that make up the genome. Adjacent to the CRISPR region were certain characteristic genes, called CRISPR-associated sequences (cas). One Cas gene that plays a particularly important role in this Narrative is called Cas9 (depicted in Figure 4). In bacterial genetics, the large majority of the DNA sequences are coding sequences (over 90% of bacterial DNA typically consists of protein-coding sequences, whereas the large majority of eukaryotic DNA is non-coding). One consequence of this more packed genome is that several genes that encode proteins with related functions are often clustered together. Thus, the co-occurrence and co-location of CRISPR arrays and cas genes strongly suggested that they are part of a unified systems with a related biological function. Thus, elucidating the function of one could give clues into the function of the other. But what was their biological function?

DNA sequences are most easily studied using computers, which can decipher information in long strings of As, Ts, Cs and Gs (see Video 1 Whiteboard on Bioinformatics). For instance, are particular sequences coding (i.e., do they encode proteins per the Central Dogma of biology), or not? Are similar sequences found in different organisms or are the sequences unique? Is the sequence of As, Ts, Cs, and Gs random, or do they form some sort of a pattern?
Eugene Koonin and Kira Makarova at NCBI (the National Center for Biotechnology Information, within the NIH) were masters at deciphering patterns in DNA sequences. They attempted several times to ascribe functions to the Cas proteins associated with CRISPR loci (failure is part of science, and this is why multiple attempts at searching turn into "re-search"). Eventually, they established that Cas proteins are related to other known proteins that interact with DNA and RNA molecules. In particular, they had some distantly related features in common with proteins involved in a defense system known as RNA interference that degrades the RNA of invading viruses. This clue led them to hypothesize that the Cas proteins may be part of an ancient prokaryotic defense system, which we will see soon is correct. However, the CRISPR-Cas defense system is different from RNA interference and indeed unlike anything that was previously known or discovered.
Clue 3: CRISPR "spacer" sequences show homology to phage DNA
What about the "spacer" sequences within the CRISPR array, the seemingly random sequences between the CRISPR repeats? The most basic computational analysis is a homology search: does a string of unknown or mysterious nucleotide sequences match something known and previously characterized genetically or functionally. Such a match might provide a powerful clue as to the function of the unknown sequence. Did the spacer sequences match any other sequences in DNA databases? Several scientists found matches, and they were surprising ones indeed. The spacers showed homology to DNA sequences from bacteriophages and plasmids, invasive elements of bacteria. This clue raised a new element of intrigue: why would there be viral sequences between repeated bacterial DNA elements? The answer came together in the Discovery, which is featured next.
The Discovery: CRISPR-Cas is a bacterial immune system
I was part of a team at Danisco (a DuPont company) working on Streptococcus thermophilus, a bacterium globally formulated as a starter culture for the industrial fermentation of milk into yoghurt and cheese. Back in the early 2000s, DNA sequencing was used to determine the complete genomic sequence of many medically and industrially important bacteria. My first job at Danisco was to sequence and assemble the genomes of bacteria used as probiotics and starter cultures in food fermentations.
I remember encountering the CRISPR repeat sequences in the early 2000s when we were sequencing the complete genome of a popular commercial probiotic strain used in yoghurt fermentation called Lactobacillus acidophilus NCFM. The repeating structure of the CRISPR locus was so mesmerizing and genetically peculiar. We had no idea what they did, but because we encountered them so frequently in genomic sequences, our instinct was that they must be doing something important. Together with my Danisco colleagues, notably Philippe Horvath, we decided to launch a research project aimed at solving the mystery of CRISPR.
As is often the case in science, we did not solve the CRISPR puzzle in one go, but we were able to bootstrap our way to an eventual answer. We got there in two steps: the first was to analyze CRISPR sequences from a wealth of genomic data that we had at our company, which led us to a hypothesis. The second step involved performing experiments to test our hypothesis. In addition to reading, you can also hear my description of the discovery in Video 2.
Step 1: Clues from analyzing DNA sequences of the CRISPR locus
Rapid evolution of the CRISPR locus
Philippe and I quickly discovered that the CRISPR sequences were highly variable, even between different strains of the same species (a strain is a subtype of a species with a distinct genetic background). This hypervariability of CRISPR spacer sequences proved useful, as we were able to use them as characteristic identifiers of different strains of S. thermophilus. In fact, genetic subtype screening is the basis for the very first CRISPR patent that was ever submitted, in 2004. Eventually, we determined the CRISPR sequences of thousands of S. thermophilus strains isolated across the globe and used for worldwide fermentation of milk into yoghurt and cheese. We now had a wealth of DNA sequence data for the CRISPR locus at our disposal. Might these data reveal the function of CRISPR?
During our CRISPR subtype analysis, we noticed something remarkable, which steered us on the right track. Fortunately, Danisco kept excellent records of numerous S. thermophilus strains over decades. Looking at a diverse collection of strains, we could group families of strains based on similar CRISPR DNA sequences and cross-reference this information with their phage sensitivity. Remarkably, certain strains of S. thermophilus, grouped by us solely based upon the DNA sequence of their CRISPR locus, were previously classified by Danisco as being particularly resistant to certain bacteriophages. Conversely, strains with other CRISPR sequences were classified as being more phage sensitive.
Explorer’s Question: What might this result suggest to you?
Answer: CRISPR is a DNA sequence in search of a function. Remarkably, this result showed a correlation between bacterial strains with similar spacer sequences in the CRISPR loci and their sensitivities to particular phage. This result correlated a genotype (a particular characteristic of the CRISPR DNA sequence) to a phenotype - whether the bacteria were resistant or susceptible to phage attack. This is not proof that the two are connected, but provides a hint that they might be.
We also noticed a second surprising result: the CRISPR locus could change over time. Danisco not only kept records of different subtypes of S. thermophilus, but they also kept records of individual strains over time. We had specific examples of a particular bacterial strain that was sampled and sequenced several years apart and could compare the sequences. While the vast majority of the genome remained unchanged, we noticed that the number of repeats in a CRISPR array could expand within a given strain even over months (Figure 5). Evolution usually sculpts changes in the genome through the relatively slow process of spontaneous mutations and natural selection (see Narrative by Koshland on Mutations). Our results, however, suggested that the CRISPR locus was evolving and changing much more rapidly than the rest of the bacterial genome. The CRISPR locus was not a static genetic feature; rather, new repeats and spacers could be added on top of those that existed in the ancestral strain. Where did these sequences come from?
The CRISPR locus has homology to viral sequences
At the same time and independently of Koonin and Mojica (see their work described in Clue 3), we used bioinformatics to look for meaningful homologies of CRISPR sequences with other sequences that had been deposited in DNA databases. Besides the obvious matches of CRISPR repeats with one another, we consistently observed imperfect but believable matches between the spacer sequences and eukaryotic viral sequences, such as monkeypox, hepatitis, influenza, and even HIV.
Why would there be eukaryotic viral sequences in the CRISPR locus? Was this contamination or an artefact? We had little reason to believe that eukaryotic viruses were infecting bacteria. Still, these homologies were not random "noise," because the likelihood of only picking up homologies with viral sequences from a large DNA database (filled mostly with prokaryotic and eukaryotic sequences) was very low if it was a totally random result.
Still, the homology with eukaryotic viruses did not make sense. However, this initial finding indicated to us that we should not solely focus on the repeats, the obvious DNA sequence elements that actually defined "CRISPR." Instead, we needed to pay attention to the spacer sequences between the repeats, which at the time were thought to be less important or wholly unimportant. As Pasteur once said, "chance favors the prepared mind," and we were now ready to pay attention.
A match between CRISPR "spacer" DNA sequences and phage that attack them
As mentioned previously, bacteria, including those used in food manufacturing, are subject to phage destruction. Accordingly, while we were sequencing S. thermophilus strains, the Danisco team was also busily sequencing the genomes of the phages found in our manufacturing environments. We were fortunate to have both sets of sequence data available within our company. With more pieces and clues available, the puzzle was starting to take shape.
By sequencing CRISPR loci of our commercial bacterial strains and the viral sequences of the phages that attacked them, we came across an extraordinary finding that was a turning point in the project. The CRISPR spacers were sometimes perfect nucleotide matches to the phage sequences that attacked them! The expansion of the CRISPR locus actually occurred by addition of DNA sequences matching those of phages. Although circumstantial, and not evidence per se, this was a rather compelling observation linking the CRISPR loci to phage infection.
To build this connection further, we had the amazing fortune of having frozen stocks of historical strains within the Danisco collection that had been isolated and characterized over decades. We unearthed strain derivatives that had been exposed to industrial phages. In some of those cases, we observed that the CRISPR loci had expanded through the addition of spacers with an exact nucleotide match to sequences found in the genome of the infecting phage that plagued our industrial plants. In some notable cases, we were able to observe larger CRISPR loci in phage-resistant bacteria that had been generated by our company, sometimes decades ago. Thus, the acquisition of a CRISPR spacer with a match to a particular phage was correlated with the ability of that bacterial strain to resist death by the phage. The plot thickened.

When we realized this, we had a strong suspicion that CRISPR was linked to immunity. Immunity refers to the ability of an organism to resist infection. Mammals, for example, have the ability to store a memory of a prior attack by a pathogen and then respond swiftly to a subsequent attack and defeat the invader. This ability to remember and respond with a counter-attack is called "adaptive immunity," which is mediated by specialized white blood cells called T cells and B cells. Adaptive immunity is the basis of vaccination, which involves an intentional exposure to an inactivated pathogen to create a stored memory for a possible future infection. Based upon our clues that (1) CRISPR loci evolved over time, and (2) that CRISPR spacers seemed derived from phage genomes, we hypothesized that CRISPR served as an adaptive immune system for bacteria and constituted a genetic vaccination record of infections.
Step 2: Experiments to test whether CRISPR serves as an adaptive immune system
After the clues came together, our next step was to devise experiments that would enable us to test the link between CRISPR loci and phage resistance. Of course, as scientists working in industry, we first filed for a patent application (time-stamped August 2005). Once a patent is filed, inventors have 12 months to reduce their invention to practice. Not only were we tackling a challenging problem, but we were racing against this 1-year clock to convert the patent with evidence obtained from experiments.
We developed three sets of experiments to demonstrate the connection between CRISPR and resistance to phage infection.
1. CRISPR loci acquire phage sequences
In the first series of experiments, we aimed to test the hypothesis that the CRISPR locus expands after the phage exposure and accompanies development of resistance. We exposed a S. thermophilus strain to a phage and let nature run its course. Experimentally, this was done by exposing a bacterial broth culture to a solution of phages and then distributing it on an agar plate (Figure 7). Expectedly, the majority of the bacterial population succumbed to the phage and died. However, an occasional robust bacterium could build resistance against phage; this small proportion of the bacterial population (a couple in a million) grew despite the presence of the virus.
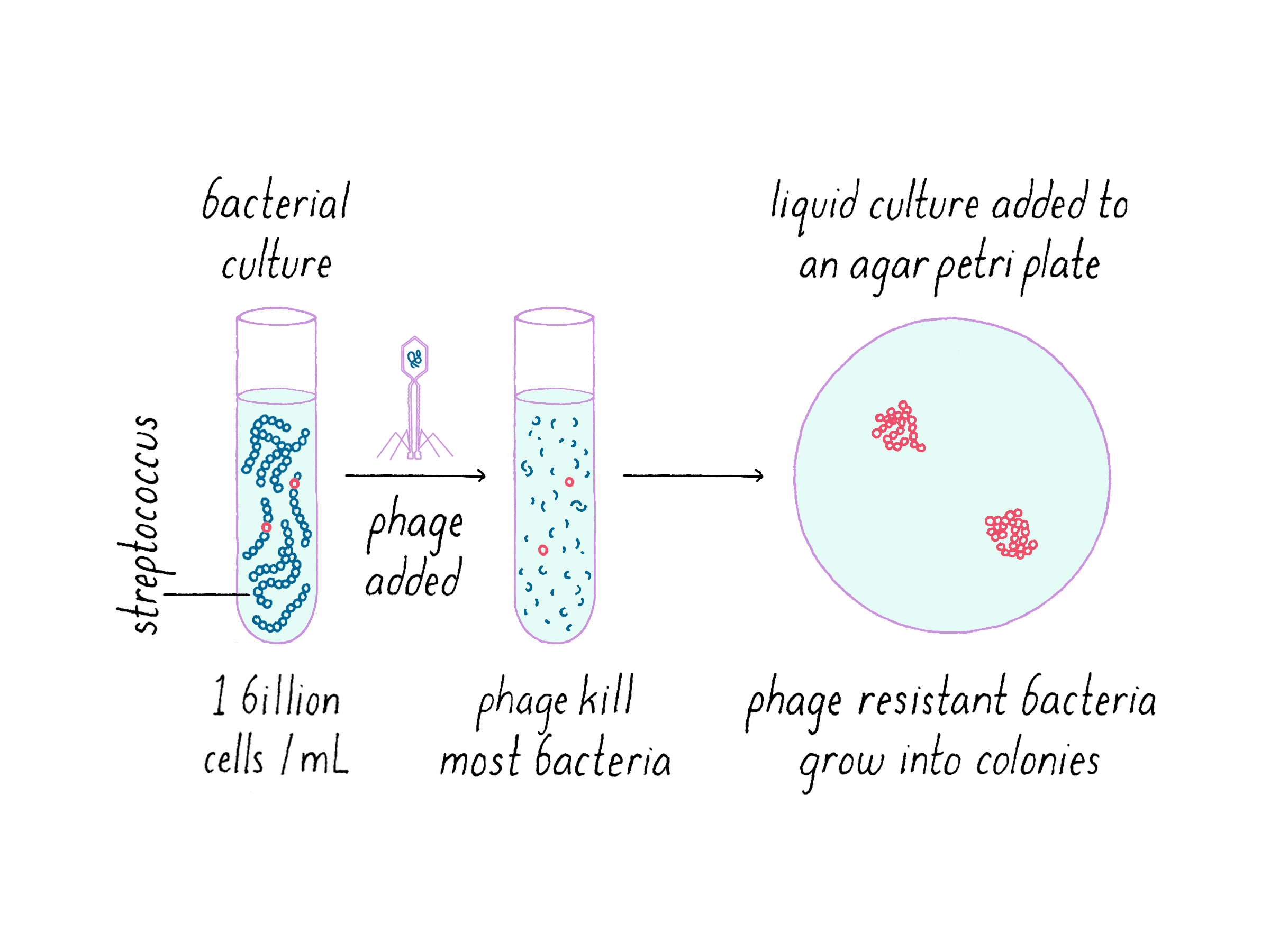
Resistance can arise by spontaneous mutations in the bacterial genome; for example, in a bacterial protein to which the phage needs to attach in order to inject its genome into the bacteria. Indeed, resistance through spontaneous mutations was the basis of the Nobel Prize winning experiment by Salvador Luria and Max Delbruck, which is described in the Narrative on Mutations by Koshland. However, in addition to spontaneous mutations, we hypothesized that some phage may become resistant through CRISPR immunity. (For discussion of various strategies of how bacteria can become resistant to phage, see Dig Deeper 1.)
In our experiment, we selected phage-resistant variants (colonies of bacteria that survived on the plate), amplified the DNA in their CRISPR loci by a method called polymerase chain reaction (PCR) and sequenced the DNA (Figure 8). Generally, the bacteria became resistant to one phage strain to which they were exposed but did not become resistant to another closely related phage that they had not seen. On the bar graph below, a value of 1 indicates a maximal "phage kill" of the original bacterial strain; smaller numbers indicate less-effective killing by the phage (note that this is logarithmic plot that decreases by powers of ten). Thus, as shown in Figure 8, this bacterial colony that was exposed to a phage A became resistant to phage A but not phage B. This was a heritable trait that the original resistant bacteria passed on to their progeny.
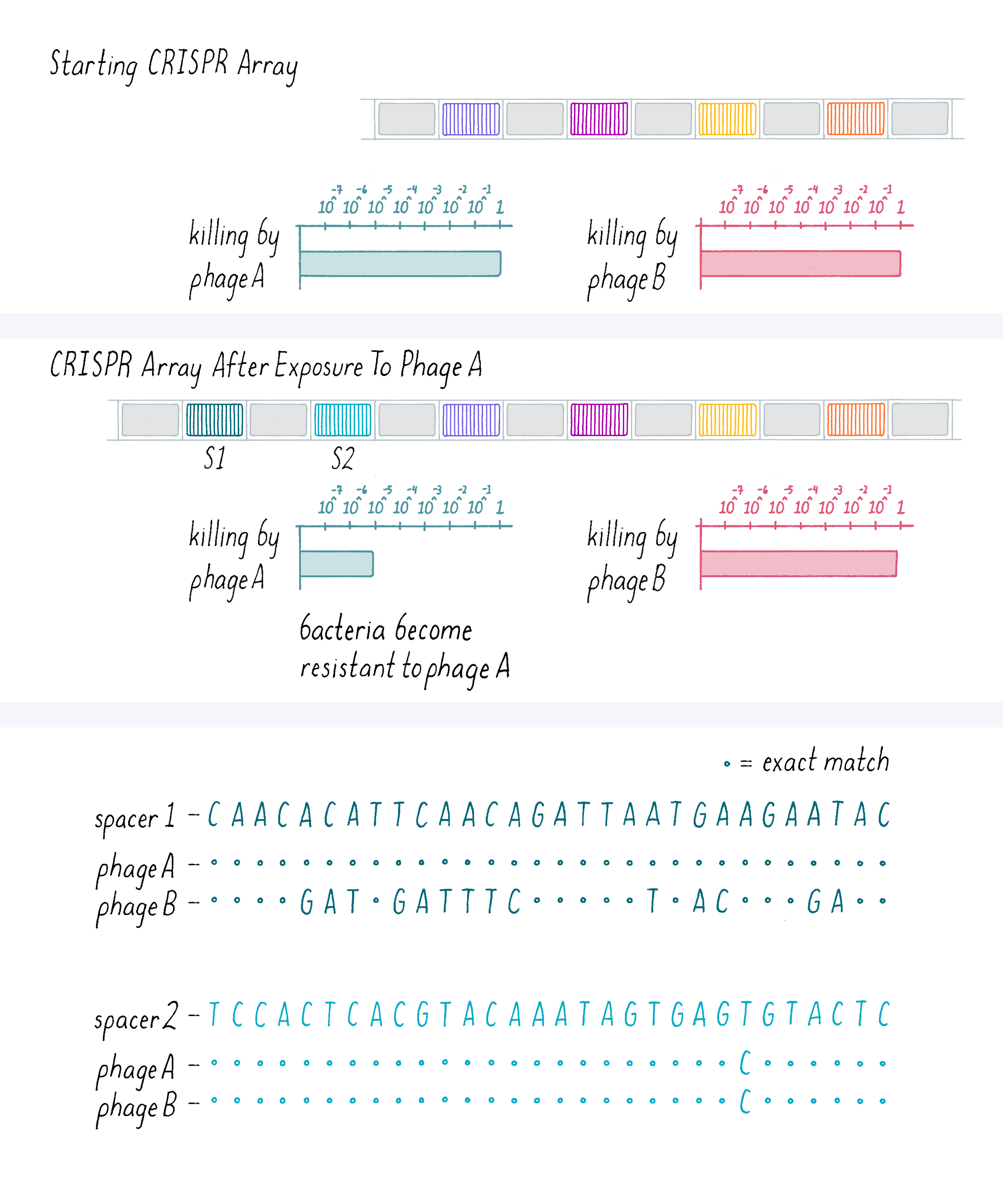
Explorer’s Question: This bacteria colony acquired two spacers. A comparison of the bacterial spacer sequences to phage A and B genomic sequences is shown above. What do you conclude about the nucleotide specificity required for phage immunity?
Answer: Spacer 1 is an exact nucleotide base match to phage A, but a poor match to phage B. Spacer 2 has a single nucleotide mismatch with both phage A and B. From these data and the information that this bacterium is not resistant to phage B, you might conclude that spacer has to be an exact match to a phage sequence in order to confer immunity. In addition to this one experiment, you would want to see this finding substantiated with more examples. Additional results have shown that some mismatches can be tolerated at the distal end only.
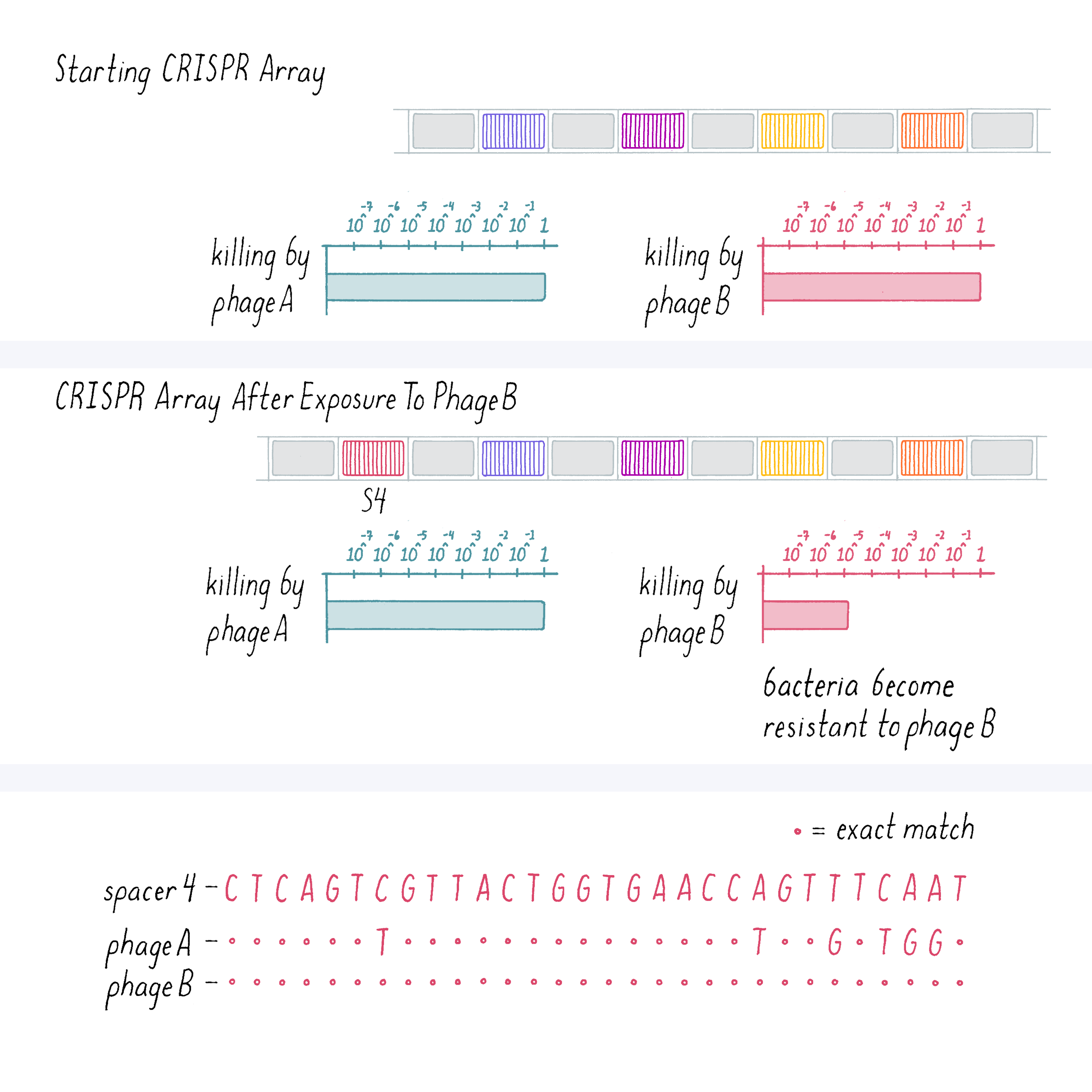
We also performed the converse experiment of infecting bacteria with phage B and then looked at the spacer sequence of the bacteria that developed resistance. In the result shown below (Figure 9), a new spacer was acquired by the bacteria that showed an exact match to phage B DNA but not phage A. And those bacteria were selectively resistant to phage B.
We also repeated the experiment with a phage cocktail of phage A and B (Figure 10). In this case, bacteria acquired immunity to both phage. The bacterial colony that we selected acquired two spacers that were perfect matches to both phage strains (strains A and B are very similar and share regions of identical sequences as well as other regions with minor differences, as in the examples shown above).

Explorer’s Question: The above resistant bacteria acquired two new spacers. What do you think would happen if one of the two spacers was deleted?
Answer: Each spacer shows sequence identity to both phages. So one spacer should be sufficient to confer resistance to both phages. However, having two spacers might make resistance more efficient.
In summary, the experiments we performed showed that bacteria that survive exposure to phage acquire novel CRISPR spacer sequences that match the antagonistic viral sequences.
Explorer’s Question: Are the above data sufficient to validate the hypothesis that the CRISPR locus provides immunity to phage?
Answer: These data SUGGEST that there may be a link between the CRISPR locus (the CRISPR genotype) and phage resistance (the antiviral phenotype). The experiment DOES NOT PROVE the link. It shows that phage infection leads to resistance AND acquisition of a new, matching spacer, but not that phage infection leads to resistance BECAUSE of the acquisition of the spacer. While we had an exciting scientific piece of data in the story, we still had to demonstrate the direct causal link between CRISPR content and viral resistance.
2. CRISPR sequences confer phage resistance
In the second series of experiments, we genetically altered the CRISPR spacer content ourselves by genetic engineering, without phage infection. We then could ask the question- if we introduced a new CRISPR spacer corresponding to a particular phage, would that genetically engineered bacterium become "immune" to that phage? This experiment would test whether there is direct link between the CRISPR spacer content and the sensitivity of the strain to viruses.
Conveniently, during in my PhD studies, I learned how to alter the genetic content of lactic acid bacteria (like Streptococcus thermophilus), and had access to the required genetic engineering tools, which were not readily available at the time. Thus, I was rapidly able to engineer the CRISPR locus by adding, removing, or swapping "spacer" regions. We showed that by:
A) adding a spacer: phage resistance was acquired;
B) removing a spacer: resistance was lost;
C) swapping CRISPR content between two strains: their respective phage resistance was switched.
Explorer’s Question: Which of the above experiments would be considered a "loss of function" and which a "gain of function"?
Answer: Experiment B is a "deletion" that would result in a "loss-of-function." Experiments A and C are "gain of function" experiments. In both cases, the genetic engineering results in the acquisition of a new phenotype of function: resistance to phage.
An example of the data from experiment C is shown below. In this case, we took the bacterium that acquired resistance to phage B and swapped out its spacers for the ones that were associated with resistance to phage A. When we did the spacer swap and retested for phage sensitivity, indeed we found that phage resistance was now switched to phage A (Figure 11).

3. Cas genes are involved in acquisition and interference
As previously mentioned in Clue 2, CRISPR loci are typically in close proximity to cas genes, and genetic association typically correlates with functional association. Given the association, we next set out to determine whether cas genes were involved in phage resistance. We used a molecular biology technique called a "knock out," which inactivates a gene by inserting a piece of DNA that disrupts the coding sequence, thereby precluding the translation of a functional protein.
We found that the two largest cas genes in S. thermophilus, cas9 and csn2, are essential for CRISPR function, but for different reasons. By inactivating csn2, the CRISPR locus was unable to acquire new spacers from phage, implicating this gene in the vaccination process. By inactivating cas9, CRISPR sequences were acquired but the bacteria were unable to resist phage attack, implicating this gene in the immunity process (Figure 12). These results show that different cas genes are involved in spacer acquisition and subsequent attack on phage invaders. The largest gene, cas9, would encode what turned out to be the most revolutionary molecular tool in a generation, as we will discuss later. While the involvement of CRISPR spacers in phage resistance had been posited by others, their reliance on associated cas proteins was unknown at the time.

What Happened Next?
We were quite excited about these results. We thought that we had definitive proof of a sophisticated adaptive immune system in bacteria. We used our three experiments described above to support our patent application. However, it took Philippe and myself 6 months to convince the management at Danisco that our results were worth publishing. It also was not trivial to convince Science magazine and its reviewers to have interest in publishing a study on a peculiar genetic locus from a yoghurt starter culture. But eventually, the paper was submitted in 2006 and published in 2007 (see Dig Deeper 2 for more information on this story of publishing the work).
While our study showed that CRISPR-Cas functions as an adaptive immune system, it did not explain how it works. In 2008, considerable progress was made when two groups characterized the mechanism of action of the CRISPR-Cas immune system. First, van der Oost and colleagues showed that CRISPR immunity is driven by RNAs (called crRNAs) that are transcribed from the CRISPR locus; each contains one spacer, thus defining a particular earlier invader of that bacterial strain. This established that CRISPR immunity is RNA-mediated. Second, Marraffini and Sontheimer showed that the phage DNA is the target of CRISPR interference. After relatively slow progress since the first observation in 1987, in a relatively short timeframe it was established that CRISPR is a DNA-encoded, RNA-mediated, DNA-targeting immune system.
Attention also turned to Cas9 and the mechanism of this important enzyme came into focus. We, in collaboration with the Moineau laboratory in Canada, showed in 2010 that Cas9 is an endonuclease that cleaves plasmid and phage DNA during infection. Equipped with this scientific understanding of the CRISPR-Cas molecular machinery, the Doudna and Charpentier laboratories repurposed Cas9 to become a versatile genome cleavage tool in vitro in 2012 (see CRISPR-Cas9 Key Experiment by Doudna et al.). The following year, multiple labs concurrently showed in human and bacterial cells that Cas9 could be programmed to selectively target DNA sequences of interest and drive genome editing by the endogenous DNA repair machinery. Although multiple groups were performing similar studies concurrently, teams lead by Feng Zhang and George Church would be the first to publish back-to-back papers in Science early in 2013, showing that Cas9 nucleases could generate efficient genome editing in human cells. This revolutionized the genome editing field and triggered the CRISPR craze that has taken hold ever-since.
Shortly after the initial demonstration that CRISPR enabled genome editing, Cas9-based editing was extended to many different cells and many entire living organisms, resulting in perhaps the most explosive new biotechnology tool to come along since DNA cloning and DNA sequencing. With DNA sequencing tools, scientists could "read" the genetic blueprint (the genome) of any organism. With CRISPR-Cas, scientists now have a tool to "rewrite" the genome. We will explore some applications of this new technology in the Frontiers section.
The growth of the CRISPR field has been amazing to witness. In 2008, we held the first scientific conference on CRISPR-Cas systems at UC Berkeley (organized by Jill Banfield and myself). The meeting was small, because only about two dozen individuals in the whole world had any interest in these systems. There were relatively few papers published on CRISPR in the scientific literature. However, the meeting started a series of field-changing collaborative efforts, and life-altering exposures to CRISPR for investigators, students, and post docs, many of whom are now world-famous scientists. Now, there are hundreds of thousands of scientists using CRISPR-based technologies in academia and industry and thousands of papers involving CRISPR being published every year. Starting from humble beginnings – curiosity about patterns of nucleotides in the bacterial genome – CRISPR is now revolutionizing all of biology
Part II: Knowledge Overview —
CRISPR-Cas systems
Bacteria occur broadly in nature and are nearly ubiquitously present in various habitats and environmental niches on the planet. Recent microbiome studies have unearthed amazing bacterial diversity in mixed microbial populations in water, soil, plants, animals, and humans. Bacteria are also widely used in the manufacturing of fermented food and have been domesticated for food preservation processes for millennia. Likewise, the viruses that infect bacteria, called bacteriophage or phage, have now been studied for over one hundred years and have been detected throughout the globe. Bacteriophage are believed to be the most abundant and the most diverse biological entity on the planet. Yet bacteria thrive, despite the ubiquitous presence of predatory viruses. How these two biological enemies, phages and their bacterial host, engage in a sustainable arms race is a fascinating puzzle. In the first part of the Knowledge Overview, I will describe our current view of how bacteria use the CRISPR-Cas9 immune system to defend themselves against phage at the level of "genomic warfare". In the second part, I will describe how this system has been re-purposed for genome engineering.
How CRISPR immunity works
Generally, CRISPR-Cas systems are composed of a CRISPR array associated with cas genes (see Journey to Discovery). The CRISPR array consists of a series of conserved CRISPR repeats interspersed with spacers, sequences from invasive genetic elements such as bacteriophages and plasmids that the bacteria acquired over time to constitute a genetic "vaccination card." If the bacteria encounter foreign DNA that matches the sequence of one of their spacers, for example, they are attacked by a phage that they have encountered previously, the spacer will direct one of the Cas proteins to cut the foreign DNA and halt the attack.
Bacteria have evolved many different flavors of CRISPR-based defense systems, reflecting the diversity of both bacteria and the viruses that prey upon them, and their widespread occurrence and distribution reflects the nearly ubiquitous presence of viruses on our planet. Discussion of other CRISPR-Cas systems can be found in Dig Deeper 3. Here, I will focus on the CRISPR system that has been most widely repurposed for genome engineering, from the bacteria Streptococcus, for which the Cas protein that cuts the foreign DNA is called Cas9.
The three steps of CRISPR immunity are discussed below: (1) acquisition, (2) expression, and (3) interference.
Acquisition
In this first step, a segment of the invasive DNA is copied and fused to a new CRISPR repeat at the terminal end of the CRISPR array by a series of bacterial enzymes (Figure 13). This allows uni-directional addition of novel spacers iteratively over time. The series of spacers captured in the CRISPR array thus constitutes a historical genetic record of the infection. By capturing a piece of the invasive DNA into the CRISPR locus, this dynamic system is adaptive (evolution in response to an environmental stimulus, such as a viral attack). It is also heritable, since once integrated into the bacterial DNA, the incorporated viral sequence will be subsequently passed on to the next generation of bacteria once the cell divides. Vaccination is not only effective against the original virus but also any related variant which contains the captured sequence, providing opportunities to build immunity against families of viruses when the acquired sequence is conserved across a group of phages.
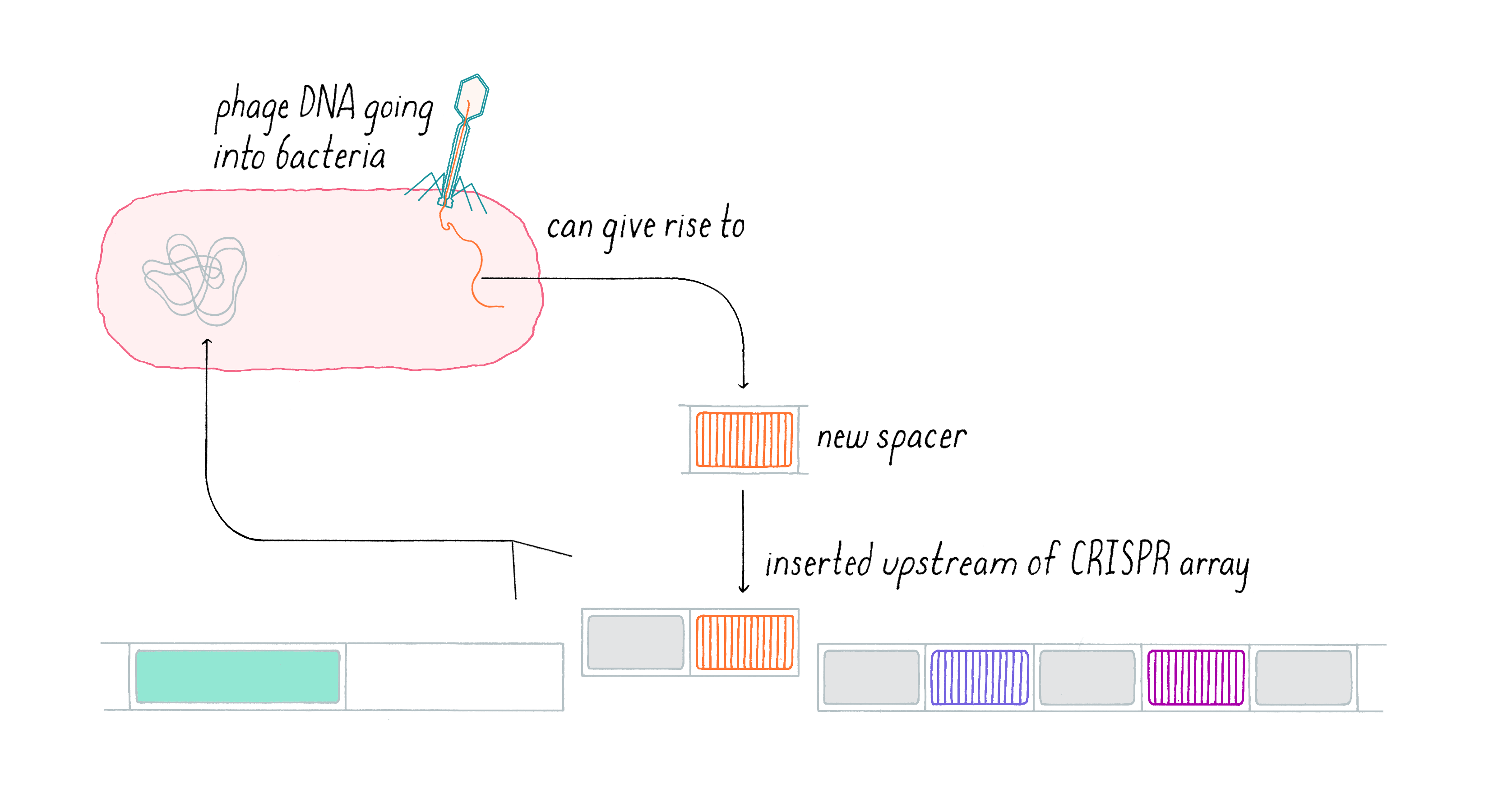
Expression
In the expression process, the CRISPR array is transcribed into a precursor CRISPR RNA molecule (pre-crRNA) (Figure 14). The pre-crRNA is then cleaved and processed into a series of small CRISPR RNAs (crRNAs) by cleavage within the CRISPR repeat sequence. This cleavage reaction involves another RNA, called tracrRNA, and the Cas9 enzyme. Each crRNA that is produced contains the sequence derived from one spacer. The spacer sequence acts as a "search" query to detect potential invasive elements that match prior infection events and allows the host to trigger the immune response when a match is found.
Since viral infections can take over the host to promote virus replication within minutes, the cell must respond very rapidly to the infection if it is to survive. Therefore, CRISPR spacers and Cas proteins are always expressed so that the cell can mount a timely immune response. However, the transcription of the CRISPR components can also be elevated after a viral attack, which sounds the alarm and generates a more robust response.
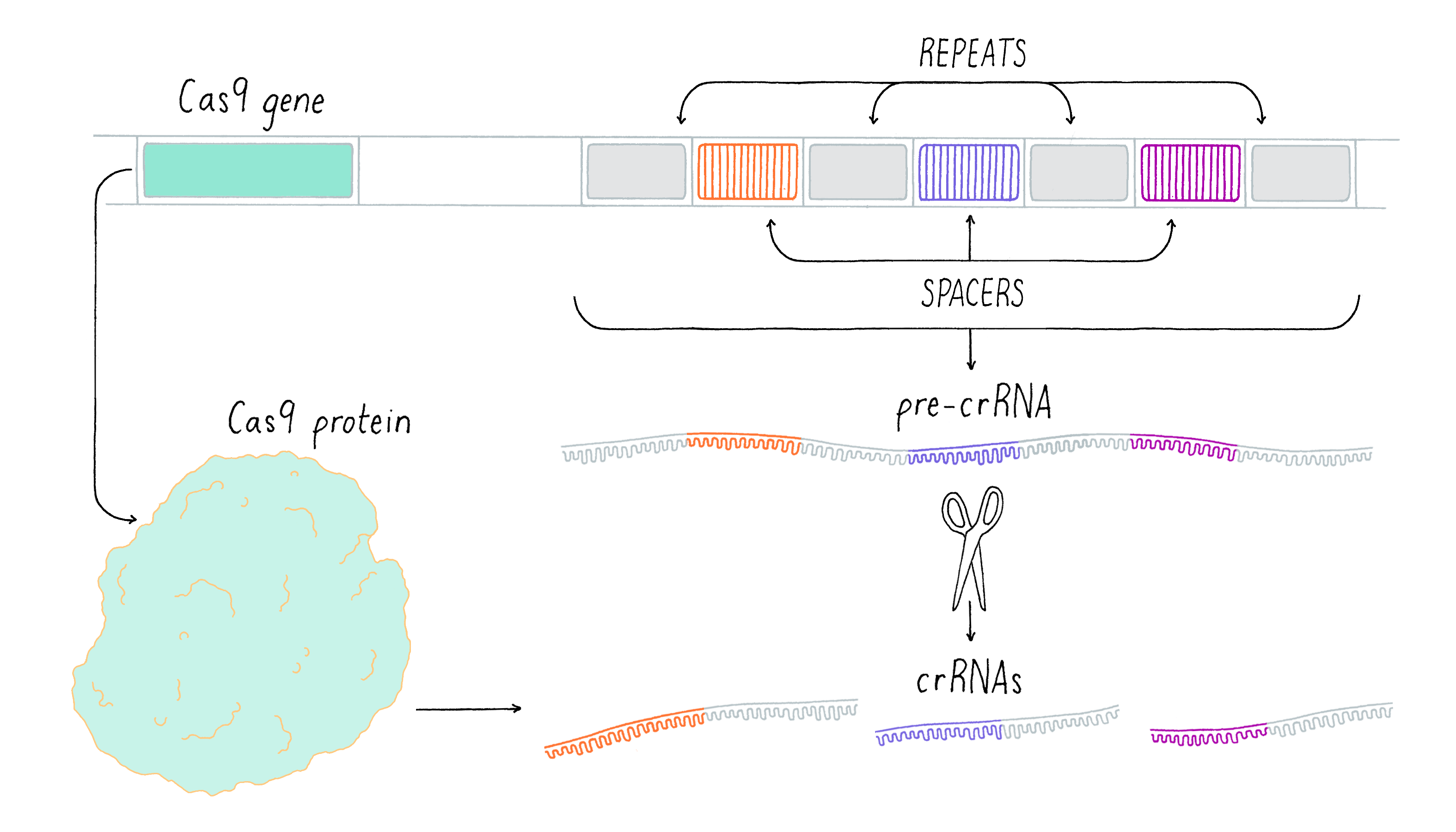
Interference
In the final step that targets the phage genome, a crRNA and a tracrRNA form a complex with the protein Cas9. The fully-loaded Cas9 then scans along DNA in a random search to find a sequence with a perfect match to the crRNA along with another more general recognition cue called a PAM sequence (see Dig Deeper 3). Cas9 scans along the DNA through a random process known as diffusion, which is discussed in the Narrative by Prakash. If a perfect sequence match to the crRNA sequence is found, then the Cas9 stops scanning and triggers an endonuclease activity; endo means "internal" (i.e. within a long DNA molecule) and nuclease is an enzymatic activity acting to sever a nucleic acid. In the case of Cas9, the enzyme cuts across both strands of the DNA helix, breaking a single long molecule into two parts (Figure 15). This single cutting activity is sufficient to foil the replication plans of a phage, which needs to preserve and propagate its genome as a single intact DNA molecule. This enables this bacterial immune system to search for sequences that match its spacer vaccination record and specifically destroy genetic elements that contain that genetic signature.

The mechanism for Cas9 targeting described above raises an interesting question. Why doesn’t CRISPR-Cas9 target the CRISPR array in the bacterial genome, because this DNA is also a perfect match for its crRNAs? The answer is that Cas9 requires a second nucleotide cue in addition to the sequence match to the crRNA. This cue is a short sequence signature (such as NGG, where N = any nucleotide) next to the spacer sequence, which is called the protospacer adjacent motif (PAM). Cas9 must first bind the PAM in the DNA, and only then can it interrogate the flanking DNA for a match for its crRNA. If there is a complete sequence match, then the Cas9 endonuclease is activated and cuts DNA. Importantly, the reliance on PAM target recognition prevents self-targeting of a bacterium’s own CRISPR array, because the spacers in the CRISPR array are flanked by the repeats, which never possess PAM sequences. This allows bacteria to distinguish self versus non-self, a key part of any immune system, and thus prevent an autoimmune response.
Repurposing Cas molecular machine
Genome editing
Genome editing involves rewriting the DNA sequence at a very specific location. For example, in the human genome, one may want to alter a single nucleotide out of three billion, for example to correct a single base pair mutation that produces sickle cell anemia (see Narrative on the Laws of Inheritance by Tilghman). The overall strategy for making an edit is to first cut the DNA close to where you want to rewrite the sequence, and then introduce a new sequence during the natural process of repairing the DNA, as will be described in this section.
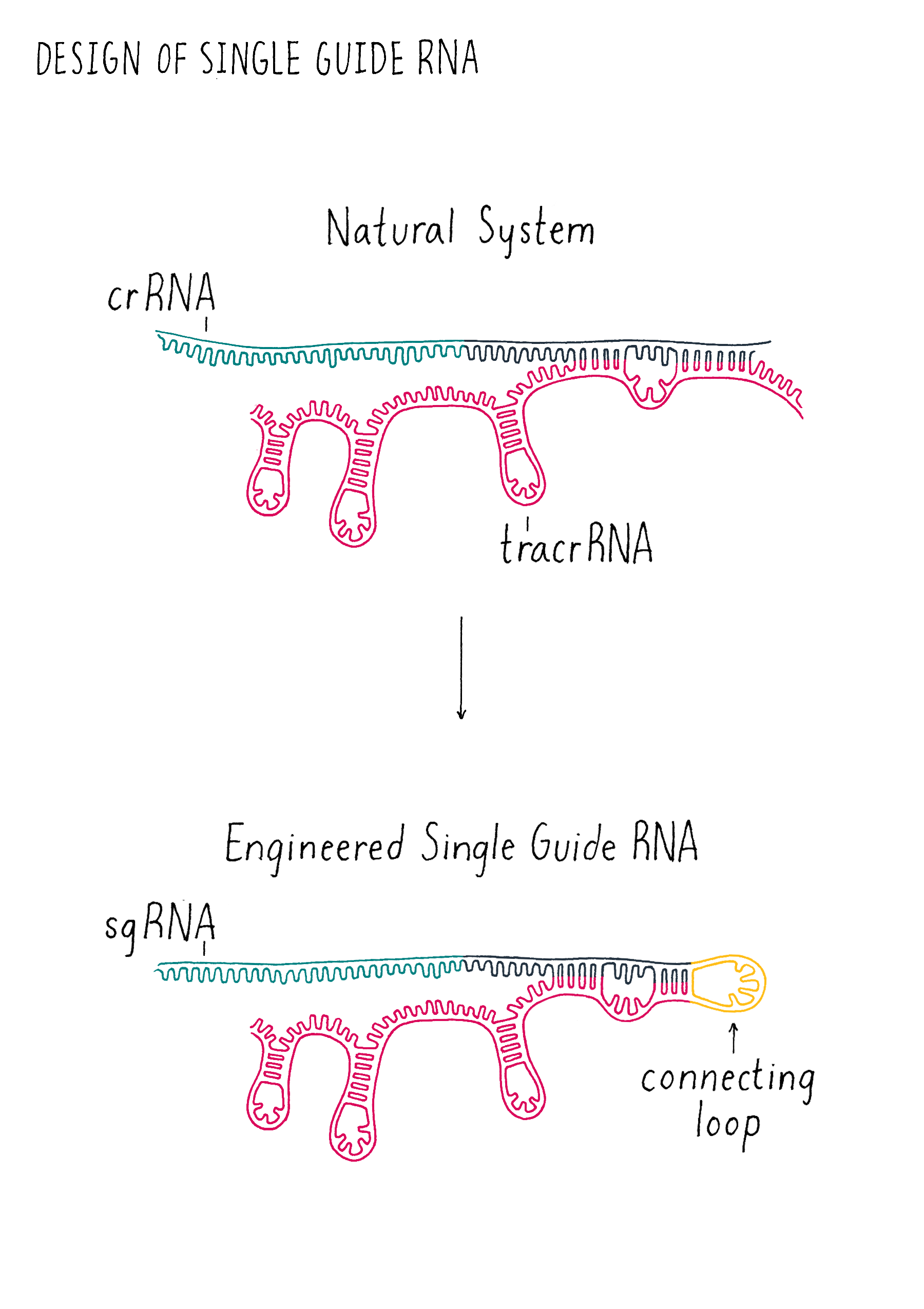
The trick is how to cut the DNA at a defined location. There were methods of cutting genomic DNA at specific locations before CRISPR. However, they involved engineering proteins to recognize and cut specific sequences. Designing and producing these specialized proteins is very slow and difficult to implement. In contrast, designing and producing specific DNA or RNA sequences is a routine process in laboratories around the world. Therefore, the world of genome editing changed in 2012 when Doudna, Charpentier and colleagues showed that the CRISPR machinery involved in the bacterial adaptive immune system could be re-engineered into a simple and programmable tool to cut DNA at virtually any defined sequence with only DNA engineering. Cas9 could be used as is; no protein engineering was necessary. They also found that the two key RNAs (tracrRNA and crRNA) involved in Cas9 cutting could be fused into a "single-guide" RNA (often abbreviated sgRNA) (see the Key Experiment by Doudna) (Figure 16), simplifying the system further. Their biochemical experiment (performed outside of cell in a test tube environment), raised the possibility that expressing Cas9 and a sgRNA in a cell would result in a specific DNA cut in a genome. Shortly thereafter, Feng Zhang, George Church, and others showed that this strategy works in a variety of cells and began generating tools to make this strategy for genome editing in cells broadly accessible.
Typically, DNA breaks are toxic and can even be lethal, especially when a cell tries to replicate its genome and divide. DNA cleavage can occur as a result of environmental insults such as radiation as well as other causes (see Narrative on Mutations by Koshland). To protect itself against such damage, cells have ways of repairing broken DNA. Once the DNA is cut, the two cleaved DNA ends can be rejoined in a process known as DNA repair (Figure 17). DNA repair is an important and complex process (see iBiology video for a more in-depth explanation), and here I will only outline the basics and particularly how it relates to DNA editing. In most organisms, there are two distinct pathways that repair double-stranded DNA breaks: non-homologous end joining (NHEJ), and homology-directed repair (HDR). NHEJ unpredictably adds a few nucleotides or generates a single base mutation, yielding a genetic abnormality at the site of cleavage and repair, analogous to the way in which repair of a cut on your skin leaves a scar. If the scar is in a non-essential, non-coding region of the genome, this quick but imprecise repair can be tolerated. But in a coding region of a gene, this can be disruptive. For example, the addition or deletion of nucleotides in numbers other than multiples of 3 would change the reading frame and disrupt the coding sequence, yielding a faulty series of amino acids and/or prematurely terminating translation and producing a truncated protein. Although such inaccuracies introduced by CRISPR-Cas9 cutting plus NHEJ repair can be deleterious for the cell, they can be very useful for scientists, by allowing researchers to study the consequences when the function of a particular gene is permanently inactivated (referred to as "knocking out" a gene or creating a knockout).
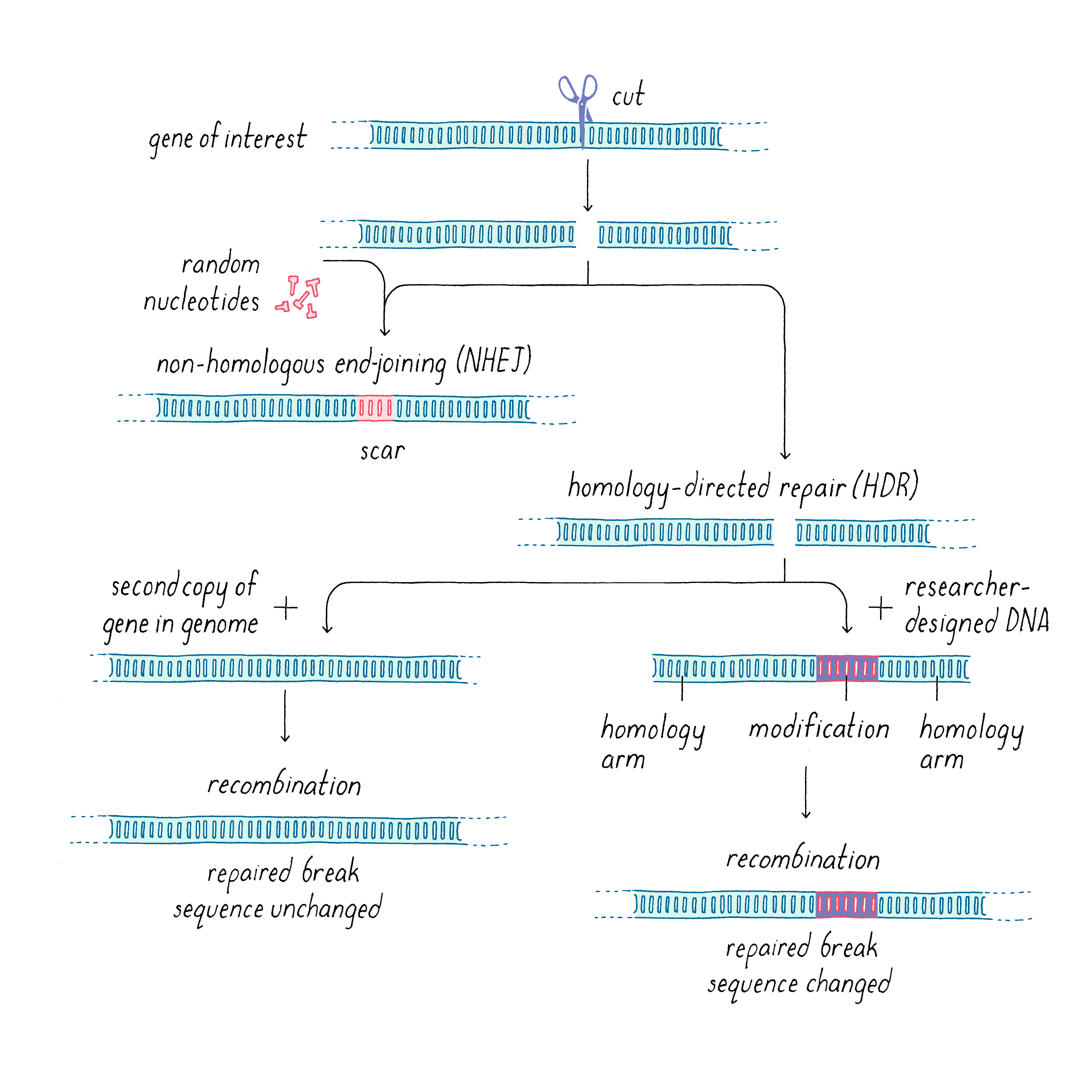
Homology-directed repair (HDR) (Figure 17), on the other hand, produces error-free repair naturally or can be deployed by scientists to introduce a well-defined edit. HDR makes use of a DNA template with a similar sequence (a similar nucleotide sequence is called a homologous sequence) to the one that was cut in order to execute the repair. Since most organisms are diploid, the natural repair template could be the second copy of the gene or potentially another similar sequence found in the genome. However, scientists can fool the repair machinery by introducing a genetically engineered repair template that is very similar to the cut gene, but also contains the scientists’ desired modification. By introducing many copies of this engineered template, it can compete with the natural template. The key for HDR is that the repair template must contain regions that are similar to the parts of the gene that flank the cut; these are called homology arms (Figure 17). These homology arms allow the template to bridge across the cut, and the damaged DNA can use the template to repair itself, introducing the engineered modification in the process. This modification can be a specific insertion, deletion, or replacement of the original sequence. The size of the edit also can be variable, from a single base pair change to a deletion or insertion of many thousands of nucleotides.
In practice, genome editing works as follows (see Figure 18):
1) First, an editing point needs to be determined by bioinformatics inspection of the gene sequence. One must find a 20-nucleotide editing site in your gene of interest with an adjacent PAM site on the 3’ end; the sequence should be unique and not found elsewhere in the genome. This sequence will be used to design a complementary sgRNA.
2) If using HDR to generate precise edits (rather than NHEJ to generate a knockout), design a repair template with homology arms adjacent to the Cas9 cut site and a central region designed to introduce an edit during HDR. Examples of edits include:
- Insert a stop codon to generate a truncated protein
- Insert a frameshift to generate a knock out
- Insert a sequence of interest (add an amino acid, a motif or a whole gene). Green fluorescent protein (see Key Experiment on GFP by Chalfie) is now frequently added to genes by this method.
- Delete a sequence of interest (remove an amino acid, a motif, or a whole gene)
- Replace a sequence of interest (change an amino acid, a motif, or a whole gene)
3) Deliver two plasmids into your cells of interest: one plasmid that will express Cas9 and the sgRNA designed in step 1 and another plasmid containing the repair template. (There are variations on this theme, but all involve delivering Cas9 protein, a sgRNA and a repair template if editing by HDR).
In summary, CRISPR-Cas9 allows the re-writing of the genome, and scientists have been successful in implementing this simple protocol in a wide range of organisms. With more effort and optimization, it may prove to be possible to edit the genome of any life form on this planet.

As wonderful as CRISPR-Cas9 genome editing is, it does not work with 100% efficiency (often more like 5–50%). Therefore, some cells receive the correct edit, while others do not. Therefore, scientists need some way to identify the cells with the correct edit (one way is to sequence the DNA at the editing site) and then isolate those cells and grow them up. Finally, there is some concern about whether CRISPR-Cas9 also cuts regions of the genome other than the intended cut site ("off-target" cuts), which can generate mutations through NHEJ. Such mutations would be of particular concern for medical applications. These off-target mutations can only be found by sequencing the entire genome. Understanding and minimizing these off-target effects is currently an important area of research.
Other uses of CRISPR-Cas9
A version of Cas9 has been engineered to lack nuclease activity, called dCas9 in which the "d" stands for "catalytically dead", or "deactivated". Thus, dCas9 can be directed by an sgRNA to bind specific DNA sequences, but not cut the underlying DNA. By fusing dCas9 to other proteins, a researcher can precisely deliver these proteins to specific segments of DNA. In this way, dCas9 fusions can be used to repress gene transcription, or activate gene transcription at specific sites in the genome. For a description of these methods, see the Key Experiment on CRISPR-Cas9 by Doudna.
Part III: Frontiers —
Uses of CRISPR-based technologies
CRISPR-Cas9 editing allows scientists around the world to re-write virtually any sequence in genomes of species across the tree of life. The method is efficient, relatively easy and relatively inexpensive, democratizing the leading edge of biology research. It is accessible to novices; many undergraduates are doing genome editing in their summer research projects. It is transforming how biological research is conducted, shaping the future of medicine, launching new companies, and eliciting a debate of what should or should not be edited (see Closing Thoughts). This is not just a new ripple in biotechnology—it is a tidal wave and we still don’t know how tall it will become.
The relative ease of implementation of genome editing in animals and plants has already led to many proof-of-concept studies that illustrate the therapeutic and agricultural potential of genome editing, a small subset of which are shown in Figure 19. In this section, I will focus primarily on applications of CRISPR-Cas9 genome editing for treating hereditary human diseases.

There are more than 6,000 genetic diseases documented to date for which a specific causal mutation in the genome has been identified. For instance, consider sickle cell disease. In this case, there is one "bad" letter (an A nucleotide is changed to a T nucleotide in sickle cells patients) in the gene encoding hemoglobin, the protein in red blood cells that carries oxygen. The altered protein also changes the shape of the cell, creating the characteristic sickle shape. Correcting the gene in the right cell type in patients might cure the disease. If the gene were corrected in an embryo, then the individual would never experience sickle cell disease in their lifetime. If the gene were corrected in the germline (egg or sperm), then that correction would be passed on permanently and become part of the gene pool.
CRISPR-Cas9 and the treatment of Duchenne Muscular Dystrophy
Let’s walk through one example of how CRISRP-Cas9 is being developed for the treatment of Duchenne Muscular Dystrophy (DMD). DMD is characterized by muscle degeneration starting in childhood and life expectancy in the teens, although some patients can live to adulthood with optimal medical care. DMD is a recessive genetic disorder caused by a mutation in a gene that lies on the X chromosome. Women have two X chromosomes. Thus, with a good copy of the DMD gene and one bad copy, they do not develop DMD and may be unaware that they are carriers of the disease. Having one X chromosome and one Y chromosome, the male offspring of a female carrier are less fortunate. If a boy inherits one bad copy of the DMD gene from mom, he will get the disease, and approximately ~1/5,000 boys are afflicted by this disease.
The genetic defect for DMD lies in a gene called dystrophin. Dystrophin plays an important role in muscle health; when dysfunctional, muscles that operate organs such as the heart and lungs fail to perform, leading to organ failure (typically heart or lung) and early death. The dystrophin gene is extraordinarily large, composed of >3,500 amino acids. In eukaryotes, most genes are divided into exons (coding amino acids) and introns (non-coding regions that are removed by splicing; see Dig Deeper 4 to learn more about RNA splicing). Dystrophin has 79 exons, one of which has a faulty nucleotide that prematurely terminates the protein. The nature of the defect in the DMD gene and the strategy to correct it is described in Dig Deeper 4.
To study human disease, scientists often create a genetic defect in an animal model that is equivalent to that found in humans. In the case of DMD, scientists have introduced the DMD mutations into mice by genetic engineering. A naturally occurring, spontaneous DMD mutation was also identified in dogs (now maintained in beagles). This dog model, in comparison to mice, shows clinical and pathological features that are similar to the human disease.
How does one change the defective gene in an animal? Here, I discuss work using CRISPR-Cas9 repair of the dog model of DMD (see the Guided Paper by Amoasii et al. in the Reference list). As we learned in the Knowledge Overview, gene correction involves delivering Cas9 and a sgRNA. To do this, scientists chose a virus for the delivery, in particular a virus called adeno-associated virus or AAV. These viruses were selected because they preferentially enter muscle cells. The genome of this virus can be engineered to include the genome editing components. For further safety, a muscle-specific promoter was used so that the Cas9 gene is expressed only in muscle cells. The therapeutic virus is grown to large quantities in a laboratory and delivered to the DMD animal by intravenous infusion or direct injection into the leg muscle tissue (Figure 20).
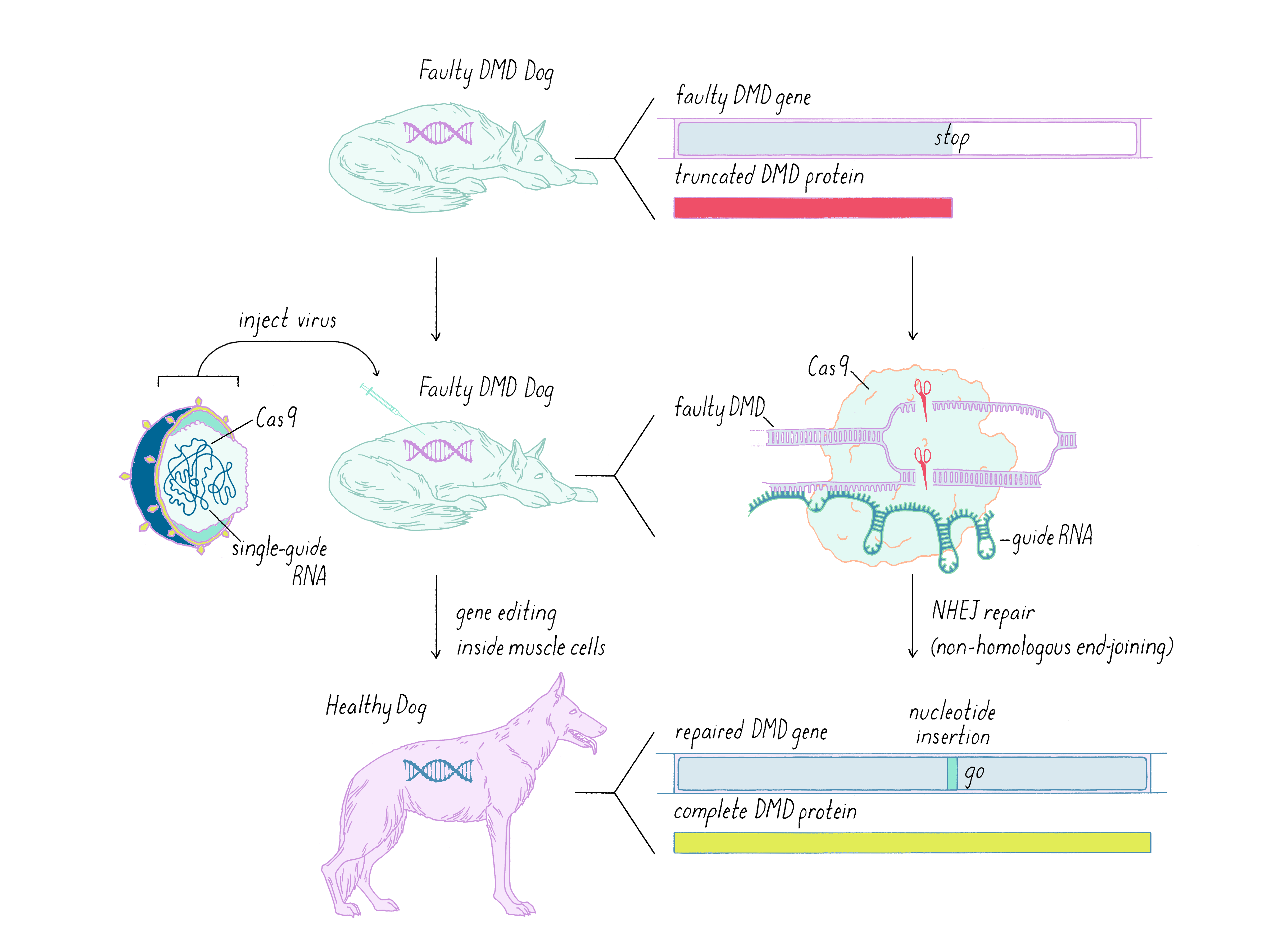
After delivering the AAV therapeutic virus and waiting 6-8 weeks, the scientists first tested whether their CRISPR-Cas9 gene editing strategy corrected the genetic defect. Not all genes were corrected, as revealed by gene sequencing. However, muscle cells are somewhat unique in containing multiple nuclei; thus, one might not need to correct all of the genes to create a functional muscle cell; perhaps one gene per muscle cell might be enough. Furthermore, some studies indicate that even the recovery of >15% of functional dystrophin could have clinical benefit for patients. Therefore, an important parameter to measure was the restoration of dystrophin protein in the DMD dogs after CRISPR-Cas9 therapy. This was tested using a technique called immunofluorescence, in which an antibody is used to probe for the presence of the dystrophin protein. In the DMD dog, the gene defect results in lack of production of dystrophin and immunofluorescence staining of muscle looks dark (Figure 21). However, when the gene is corrected by CRISPR-Cas9, then the bright staining indicates that the normal protein is now produced. The level of dystrophin recovery varied in different muscles but many muscles were restored to >50% (including >90% in the heart).
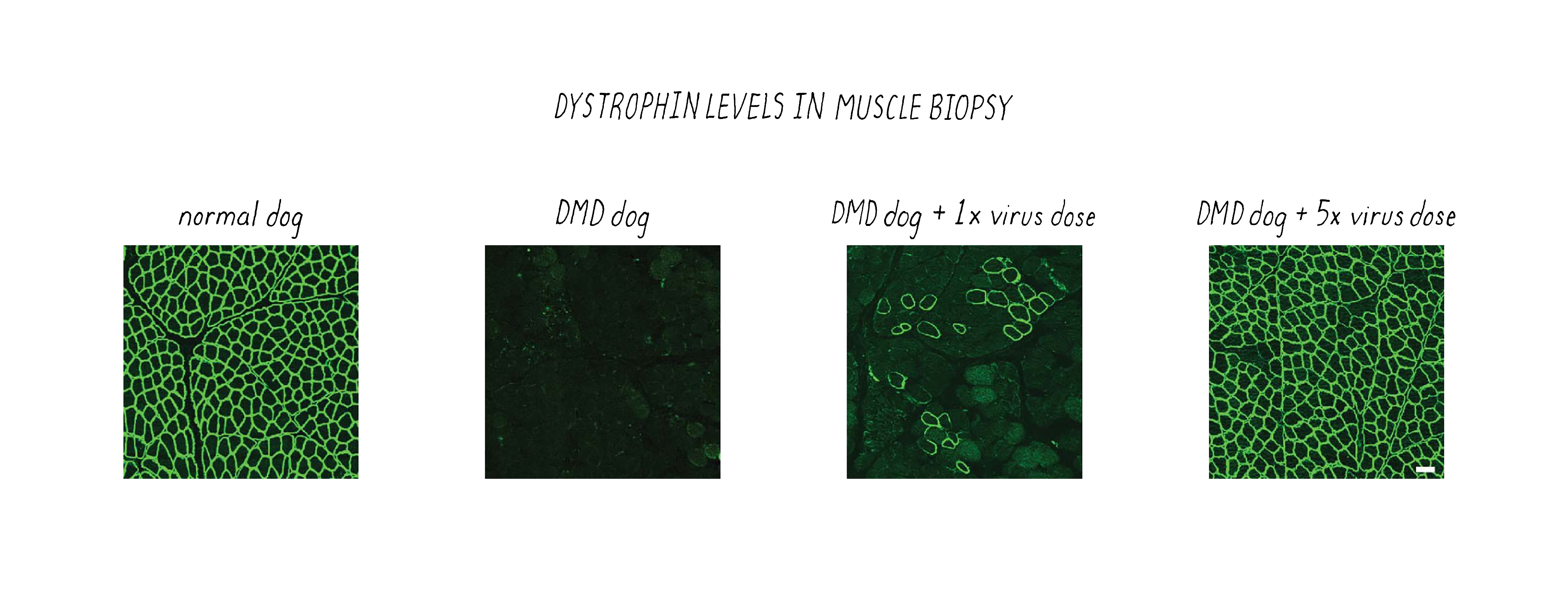
Importantly, the abnormal pathology of the muscle in the DMD dogs was restored to a healthier state by CRISPR-Cas9 therapy (Figure 22).

These results provide a promising basis for the development of engineered viruses carrying CRISPR payloads to treat patients with DMD. More generally, this study illustrates how CRISPR-based genome editing approaches can be exploited to target specific mutations associated with disease by co-opting the endogenous DNA repair machinery to correct a specific genetic defect. Many companies are working to develop CRISPR-Cas9 therapies to treat many human diseases. Stay tuned and follow the news. The first human clinical trials are happening as this Narrative was being written. This is going to be an exciting area with broad implications for human health.
Other examples of uses of CRISPR-Cas technology
Within a couple of years of the genesis of CRISPR technology, hundreds of laboratories in industry and academia have been performing genome editing of commercial plants such as corn, soybean, wheat, rice and sunflower, as well as vegetables and fruits including mushrooms, tomatoes, lettuce, strawberries, apples, and other crops (tobacco, sweet potato, cassava). As an example, using CRISPR to knock-out the gene responsible for browning in the white button mushroom, scientists have extended the shelf-life of produce. Other obvious goals include targeted genetics to increase yield, drought tolerance, water usage, pest resistance, and optimizing lipid and protein composition for increased nutrition and digestibility. Overall, these approaches can be used to optimize crops for yield and nutritional attributes, which is critical in light of our limited arable land, and the need to feed a rapidly expanding human population (see also Narrative on Plant Genetics by Ronald).
CRISRP-Cas approaches are also being used to breed next-generation livestock with desirable attributes such as the absence of horns in cows, or lowering the fat content in pigs. Recent advances in genetics and breeding for pork, dairy, beef, and poultry open new avenues for increasing resistance to diseases that afflict livestock, including African Swine Fever (ASF), Bovine Respiratory Disease (BRD), and Porcine Reproductive and Respiratory Syndrome (PRRS). For example, in the case of PRRS, efforts are underway to remove a region in a membrane receptor that allows the virus to enter the cell.
Also, this technology can be implemented in industrial bacteria, yeast, and algae used in the bioindustry for the genesis of a wide diversity of products that include commercial biomolecules, drugs, enzymes, and biofuels.
Although CRISPR-based technologies are already powerful and having significant impacts in the medical, agricultural and biotechnological industries, it is important to note that this relatively new technology is still being enhanced and optimized by many scientists around the world. Of note, major goals include enhancing the efficiency and specificity of the Cas machinery, developing strategies to deliver the CRISPR-Cas9 machinery to the desired cells and tissues, and improving our ability to predict and control DNA repair mechanisms and outcomes. There are many innovations still to come.
It is also important to keep in mind that the translation of scientific knowledge into valuable technologies with commercial applications hinges on regulatory approval and public acceptance. Thus, implementation of these new technologies must proceed with caution and also consider ethical implications. It will be critical to engage the public on these issues via an open and transparent dialogue.
Closing Thoughts
I have been privileged to participate in, along with colleagues and collaborators, some of the most exciting observations and discoveries of the early CRISPR work. I have also enjoyed witnessing CRISPR technologies being translated into actual products impacting medicine, agriculture and biotechnology. I believe that CRISPR will be viewed as one of the most impactful and disruptive technologies that have emerged in the history of the life sciences.
I am in awe of the speed and scale at which the CRISPR story has unfolded. In merely a decade, CRISPR has evolved from a study of peculiar regions in bacterial genomes, to an intriguing immune system in bacteria, and now a potent molecular machine able to readily alter the genomes of humans, other animals, plants and bacteria at will. CRISPR research is still unfolding at a frenetic pace. The global impact and rate of adoption of this technology is illustrated by the dissemination of CRISPR constructs by Addgene, a not-for-profit repository, which has shipped over 100,000 CRISPR-related plasmids to over 5,000 laboratories in over 100 countries, making this technology broadly available to all. Consequently, approximately one million scientists are currently developing next-generation gene therapies for medicine, enhancing desirable attributes of plants and livestock for agriculture, and engineering novel microbes for biotechnology. This is only the beginning of the CRISPR era.
With so much at stake and so many fields impacted by this powerful technology, however, there is a need for scientists to be mindful of the societal implications of genome editing, especially with regards to editing humans, and the germline in particular. Several scientific societies have already engaged in an open dialogue to consider the ethical implications of human germline modification and several countries have implemented a moratorium preventing these applications. Medical and scientific boundaries—guidelines and guardrails—need to be agreed upon by a diverse group of stakeholders, and account for concerns about modifying humankind and impacting future generations of individuals.
Commercial implementation of this technology in agriculture hinges on regulatory frameworks that allow the use of CRISPR for genome editing and, thus far, different regulatory agencies have differed in their approach to regulating editing technologies. Countries like the United States and Japan have elected not to regulate gene editing for agriculture, whereas the European Court of Justice has ruled that edited crops fall under the existing genetically modified organism guidelines that restrict commercialization in the European Union. These are reminders that, while scientists rapidly advance on the technological highway and open new avenues for medicine, agriculture and biotechnology, the actual development and commercialization of therapies, crops, livestock and biological products will depend on a supportive and enabling regulatory network, as well as consumer acceptance. Given the widely differing public opinions and diverse regulatory agencies operating across the globe, it is still unclear how easily and quickly these therapies and products will be made available to humankind on a global basis. With some widespread skepticism about science in general, and un-founded reservations about genetic engineering in particular, we are reminded that sometimes consumer opinions can be a difficult hurdle to overcome for powerful but disruptive technologies.
The CRISPR story that has unfolded over the past decade is also interesting from a scientific narrative standpoint. The rise of this topic from its discovery in bacteria to scientific stardom in a relatively short time-span illustrates how rapidly scientific advances can develop, fueled by interdisciplinary collaborative teams that transcend the boundaries between industry and academia. There are also many examples of how serendipity and lucky timing can drive the scientific process and propel a field forward. Finally, it is important to remember that none of the CRISPR pioneers could have predicted at the onset that studying a bacterial immune system would lead to new strategies for treating and potentially eliminating some of the most devastating human diseases. This is a theme that has emerged over and over again in the history of science—that great new technological advances start humbly with a curiosity for how life works, and then evolve through serendipitous observation and putting clues together, step-by-step.
Dig Deeper
Dig Deeper 1: Strategies for resistance to phage infection beyond CRISPR immunity
In the past few decades, advances in Microbiology and Molecular Biology have enabled scientists to observe and dissect the interplay between phage and bacteria, and characterize the defense systems that enable microbes to escape viral predation. CRISPR is one among many strategies that bacteria use to evade viral attacks. Some bacteria fight phage infection by expressing enzymes that destroy non-self DNA (this is known as a restriction-modification system). In other cases, a phage-infected bacterium may activate a program that causes it to die prematurely, thereby limiting the ability of the phage to replicate and infect other bacteria in the population. This kind of self-sacrifice to protect the rest of the population is called abortive infection. Mutations in the bacterial proteins that phage use to attach to the bacterial cell surface can also impede or block phage infection. The relative contribution of these various phage resistance mechanisms varies widely across phylogenetic groups of bacteria. In the case of Streptococcus thermophilus, the large majority of the survivors of phage infection are typically those that have acquired CRISPR-mediated immunity. This species is particularly enriched in CRISPR-Cas systems: in the thousands of strains tested globally in the dairy industry, 100% encode CRISPR-Cas systems. Remarkably, these genomes typically encode multiple CRISPR-Cas systems of different types that can all be concurrently active. Indeed, there are several reports in the literature showcasing how the CRISPR1 and CRISPR3 CRISPR-Cas systems in dairy bacteria can concurrently acquire spacers from the same phages.
The CRISPR-Cas story, in which a phage defense system was repurposed to fuel the genome editing revolution, is reminiscent of a previous chapter of the history of molecular biology. In the 1960s and 1970s, details of the restriction-modification phage defense systems mentioned above were uncovered, and repurposed as restriction enzymes to drive the recombinant DNA revolution. As we move forward and continue to mine bacterial genomes for novel CRISPR-Cas systems, we are reminded that the next “CRISPR" might be awaiting discovery. Indeed, some recent work has already shown that other novel defense systems exist, which may be the basis for the development of tools that will fuel the next revolution in biology.
Dig Deeper 2: Publishing of the Journey to Discovery paper in Science
Altogether, results from these three experiments were filed at the USPTO in the summer of 2006 to convert the aforementioned patent, and it took Philippe and myself 6 months to convince the Danisco management and the reviewers that these results were worth publishing. At the same time as we were doing our work, other scientists were independently assembling complementary pieces of the CRISPR puzzle: the paper by Mojica et al., published in 2005, revealed that CRISPR spacers show homology to viruses and plasmids; the paper by Makarova et al. in 2006 suggested that CRISPR-Cas may be bacterial immune systems. This group of papers reflects how independent groups can often take similar paths in parallel. Our Danisco team was ideally positioned to assemble this puzzle given the organism on which we worked, the presence of an active CRISPR-Cas system, the availability of genomic sequences for both the host and the viruses, the availability of historically relevant biological material, and perfect timing in the sense that some of the clues were already available to put us on the right trail, while it was obscure enough for others to remain in the darkness.
In addition to the difficulties we internally encountered in our efforts to convince the Danisco team to spend time and allocate resources to prepare a manuscript for submission to a journal like Science, we also had to convince the editors and reviewers at the journal that our work was worthy of their consideration. Initially, the Science reviewers showed relatively little interest in work on a peculiar genetic locus from a commercial yoghurt starter culture. However, yet again, luck manifested itself in the shape of timing. Actually, the delay between our initial patent filing and the original manuscript submission could not have been more fortuitously productive. Unbeknownst to us, just a couple of weeks before we submitted our paper, the famous Jill Banfield had submitted a similar story to the very same editor, at the same journal, discussing circumstantial interplay between CRISPR loci and viral sequences. She observed this interplay in metagenomic data she had analyzed from acid mine drainage environments in which bacteria co-evolved with viruses. Luckily for us, she had raised interest at the journal on this very topic at this very point in time, and I believe this is a primary reason why our paper was originally reviewed and eventually published.
Dig Deeper 3: Other CRISPR-Cas systems in bacteria
There are many diverse CRISPR-Cas systems, reflecting both the natural diversity and divergent evolution of the many bacteria that exist and have evolved over time (see Figure DD3). In all of these systems, immunity is DNA-encoded (invasive DNA is incorporated as spacers into the CRISPR array), RNA-mediated (involving the production of crRNAs) and nucleic acid-targeting (the systems specifically recognize nucleic acid sequences complementary to the crRNA).
In general, CRISPR-Cas systems are split into two main classes, six major types, and 33 sub-types that vary in their biochemical processes. For instance, class 1 systems rely on a multi-protein complex to carry out the interference process, whereas class 2 systems use a single protein to target and cleave nucleic acids. Within each class, there are different types that are each defined by a signature Cas protein, which is responsible for a specific type of nucleic acid cleavage as follows:
• Type I: the Cas3 exonuclease in complex with Cascade (CRISPR associated complex for antiviral defense) targets double-stranded DNA (dsDNA) using PAM targeting on the 5’ edge of the protospacer. The exonuclease selectively nicks and chews the target strand in a 3’ to 5’ exo-nucleolytic manner, creating extensive damage in the viral DNA. This is likely why this “Pacman-type" system is the most widespread in nature.
• Type II: the Cas9 endonuclease targets dsDNA and the RuvC and HNH nickase domains each nick one DNA strand and generate a blunt cut. This involved PAM targeting at the 3’ end of the protospacer. This system generates a clean DNA cut (a double-stranded break – DSB), which is why Cas9 is often referred to as a molecular scalpel and is useful for triggering DNA repair pathways linked to genome editing.
• Type III: the Cas10 nuclease targets single-stranded RNA (ssRNA) and together with the Csm or Cmr complex cleaves the target RNA at multiple locations. There is no dependence on a PAM sequence in this type.
• Type V: the Cas12 endonuclease nicks two strands of dsDNA at different locations to generate sticky-ends with overhangs. This involves a PAM flanking the 5’ edge of the protospacer.
• Type VI: the Cas13 nuclease nicks complementary ssRNA and also carries a non-specific RNase activity that creates “collateral damage."

Dig Deeper 4: Correcting the genetic defect of DMD
The Dystrophin gene is extraordinarily large, comprised of >3,500 amino acids. Dystrophin is divided into 79 exons with a corresponding number of intervening introns. DMD mutations disrupt the dystrophin reading frame, introducing premature stop codons that generate truncated and unstable protein. (See Dig Deeper 4 Video for a description of exon, introns and RNA splicing). There are thousands of different types of DMD mutations, but many DMD mutations ultimately cause a shift in the reading frame starting from exon 51. Therapies that restore the reading frame at, or after, exon 51 could benefit ~13% of DMD patients. The DMD dog is a model for just such a mutation: exon 50 is missing, and the resulting fusion of exon 49 to exon 51 creates a frameshift that generates a stop codon in exon 51 (see the figure).
Viral vectors were designed to target the CRISPR/Cas9 machinery to exon 51, and the resulting viruses were injected into the muscle of DMD dogs. The Cas9 molecular scalpel was guided to the exon 51 sequence, where it generated a double-strand break that recruited the DNA repair machinery. NHEJ repaired the break, which generated a variety of insertions, deletions and substitutions. Many of these did not fix the frame shift, but since the gene is already non-functional, they did not do any additional harm. However, a relatively frequent consequence of this repair was that a single nucleotide was added at the cut site. This one nucleotide insertion restored the wild-type reading frame, so that the gene could now produce functional protein to support muscle function.
References and Resources
Guided Papers
Barrangou, R., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709-1712.
This paper describes how the CRISPR-Cas system functions as an adaptive bacterial immune system. This knowledge allowed subsequent development of CRISPR-Cas as a versatile genome editing method.
DownloadAmoasli, L. et al (2018). Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362: 86-91.
The authors use CRISPR-Cas gene editing to correct the effects of mutations in the gene encoding dystrophin that cause Duchenne muscular dystrophy (DMD). This paper shows the efficacy and safety of the CRISPR-Cas system in large mammals.
DownloadReferences
-
Ishino, Y. Shinagawa, H, Makino, K., Amemura, M.
and Nakata, A. (1987). Nucleotide sequence of
the iap gene, responsible for alkaline
phosphatase isozyme conversion in Escherichia
coli, and identification of the gene product. J.
Bacteriology 169: 5429-5433
The first description of the CRISPR sequence described in Clue 1 of the Journey to Discovery.
-
Mojica, F.J., Diez-Villasenor, C,
Garcia-Martinez, J., and Soria, E. (2005).
Intervening sequences of regularly spaced
prokaryotic repeats derive from foreign genetic
elements. J. Mol Evol. 60: 174-182.
This paper shows a connection between spacer sequences in CRISPR and viral sequences, as discussed in Clue 2 of the Journey to Discovery.
-
Markarova, K.S., Grishin, N.V., Shabalina, S.A.,
Wolf, Y.I., and Koonin, E.V. (2006). A putative
RNA-interference-based immune system in
prokaryotes: computational analysis of the
predicted enzymatic machinery, functional
analogies with eukaryotic RNAi, and hypothetical
mechanism of action. Biol. Direct. 1: 7.
This paper makes a connection of the Cas genes with a possible defense mechanism for bacteria, as discussed in Clue 2 of the Journey to Discovery
Resources
-
CRISPR-Cas9 Video:
https://www.youtube.com/watch?v=2pp17E4E-O8
A short video that gives a basic explanation of how the CRISPR-Cas9 system functions in bacteria and how scientists have adapted the system to work in other organisms.
-
Q and A’s with Rodolphe Barrangou:
https://www.pnas.org/content/114/28/7183
A CRISPR centric question and answer session with Dr. Barrangou done by PNAS.
-
Dr. Barrangou Lecture:
https://vimeo.com/255061822
A video of Dr. Barrangou giving a lecture on how CRISPR was discovered, the research he has done and the implications of the technique for the future in the context of plant research.
-
CRISPR Short Video:
https://www.youtube.com/watch?v=9IgLrOEsauk
A short video featuring Dr. Barrangou explaining the CRISPR system and it’s implications.
-
CRISPR FAQ:
https://www.broadinstitute.org/what-broad/areas-focus/project-spotlight/questions-and-answers-about-crispr
A web page that includes a short video explaining the CRISPR-Cas9 system and has a series of frequently asked questions and their answers.
-
CRISPR-Cas9 Mechanisms and Applications:
https://www.hhmi.org/biointeractive/crispr-cas-9-mechanism-applications
An interactive simulation of both how the CRISPR-Cas9 system functions and what the real world applications are.
-
CRISPR Slide Show (PDF):
http://ilsi.org/wp-content/uploads/2016/07/7.-BARRANGOU-Rodolpe-CRISPR-Cas.pdf,
This is a PDF presentation slides created by Dr. Barrangou discussing CRISPR and it’s impacts on the world of food science reserach.
-
Jennifer Doudna CRISPR Explanation:
https://www.youtube.com/watch?v=SuAxDVBt7kQ
A video featuring Dr. Jennifer Doudna explaining how they discovered the CRISPR-Cas9 system, how it works and the directions it is moving today.
-
Jennifer Doudna CRISPR TED Talk:
https://www.youtube.com/watch?v=TdBAHexVYzc
A TED talk video that features Dr. Jennifer Doudna explaining the discovery, potential functions and ethical issues surrounding the CRISPR-Cas9 system.
-
Lecture on DNA repair by Jim Haber (Brandeis
University) on iBiology.org
https://www.ibiology.org/genetics-and-gene-regulation/mechanisms-dna-repair/#part-1