Quorum Sensing: How Bacteria Communicate
Bonnie Bassler
 Bonnie Bassler
Bonnie Bassler
Bonnie Bassler is a microbiologist and teacher who investigates the molecular mechanisms that bacteria use for communication and coordinating group behaviors. The process Bonnie studies is called quorum sensing. Bonnie’s research is paving the way to the development of new therapies for combating deadly bacteria by disrupting quorum-sensing-mediated communication. Bonnie is passionate about increasing diversity in the sciences. She is actively involved in educating lay people in science. Bonnie can often be found at community events, in science videos and interviews on the web, TV, and the radio, and doing policy work to make sure that everyone gets opportunities to learn and participate in science.
What’s the Big Deal?
Bacteria can communicate, and they speak multiple languages! Bacteria use chemicals as their “words.” They use chemical communication to distinguish their own species from others, and in doing so, presumably reveal friend from foe. Bacteria release their chemical communication molecules into the extracellular environment. When the level of these chemicals builds up to a critical level, a signal is relayed to the cell interior, which alerts each bacterial cell that other bacterial brethren are in the neighborhood and that they have reached a “quorum.” The entire population of bacteria then act as a large, coordinated group, carrying out tasks that would be unsuccessful if a single bacterium acted alone. This process, called “quorum sensing,” controls bacterial behaviors ranging from symbiosis to virulence to biofilm formation to natural product production. Scientists are developing cunning strategies to turn off or turn on quorum sensing on demand. Disrupting quorum sensing has the potential to stop harmful bacteria from making humans, animals, and plants sick. Conversely, promoting quorum sensing in particular beneficial bacteria could make us healthier and could lead to valuable products for medicine and industry.
Learning Overview
Big Concepts
Quorum sensing allows us to learn about signal transduction, gene regulation, cell-cell communication, and collective behaviors, all general properties of life.
Bio-Dictionary Terms Used
Antibiotic, Bacterial Colony, Central Dogma, Cytoplasm, Enzyme, Eukaryote/Eukaryotic, Gene, Gene Expression, Fractionation/Fractionate, Microbiome, Mutant, Pathogen, Plasmid, Plasma Membrane, Prokaryote, Promoter, RNA Polymerase, Symbiosis, Transcription, Transcription Factor, Virulence Factor
Terms and Concepts Explained
Autoinducer, Bioluminescence, Biofilm, Kinase, Operon, Phosphatase, Phosphorylation, Positive Feedback Loop, Quorum Sensing, Receptor, Signaling, Transposon Mutagenesis
Introduction
-
Bacteria communicate with one another, not with words, but with chemicals called autoinducers. When autoinducer levels start to increase, the bacteria know that there are many other cells around and, as a group, they start to exhibit new behaviors that are only effective when many cells act together.
-
Quorum sensing is involved in biofilm formation (communities of bacteria adhered to surfaces), pathogenesis, symbiosis, and many other processes.
Part I: Journey to Discovery - Quorum Sensing and the Molecules Involved
-
Quorum sensing was originally discovered in bacteria that could take up DNA from the environment and in marine bacteria that emit bioluminescence (make light). We will describe the two experiments that showed for the first time that bacteria communicated with chemical molecules and that they could act in groups.
-
The original findings were considered irrelevant for many years because the results were thought to be isolated to a few obscure bacteria. Later, quorum sensing was shown to be widespread in the bacterial world and crucial for bacteria that cause disease.
-
We will describe how strategies using gene transfer (gain-of-function) and genetic mutations (loss-of-function) contributed to the identification of the proteins involved in quorum sensing and to the understanding of the signal transduction mechanism.
Part II: Knowledge Overview - How bacteria talk to one another
-
Bacteria communicate using small molecules called autoinducers. The autoinducers are released by the bacteria into the environment. The higher the bacterial population density, the higher the concentration of the autoinducer in the environment.
-
Once an autoinducer has reached a threshold concentration, it can bind in sufficient amounts to a partner receptor protein that is made by the bacterium. Autoinducer binding changes the receptor’s activity (either its phosphorylation state or its ability to bind DNA). This change elicits an alteration in the expression of genes encoding proteins that enable group behaviors. The sequence of events from receptor binding to the change in gene expression is called a signaling cascade or signal transduction.
-
One gene that is typically activated by quorum sensing is the gene encoding the enzyme that produces the autoinducer itself. Thus, when the bacteria detect the buildup of autoinducer, they make even more autoinducer. This arrangement is called a “positive feedback loop,” and, in this case, feedback ensures that all nearby cells switch on their quorum-sensing-controlled genes.
-
Quorum-sensing-controlled behaviors are typically ones that require many bacteria acting in concert to make the behavior effective: bioluminescence, virulence factor production, biofilm formation, symbiosis, and release of exo-products (so-called public goods that are shared by all members of the community).
-
Bacteria often live in communities consisting of multiple bacterial species. In such environments, individual bacteria need to be able to detect how many of their own species are present versus how many other species are present.
-
To distinguish self from non-self, bacteria release multiple quorum-sensing autoinducers, some of which only one particular species can sense and some of which can be detected by a few or many bacterial species. The blend of autoinducers perceived by a bacterium corresponds to how many and what bacterial species are nearby. Decoding blends of autoinducers allows bacteria to carry out appropriate quorum-sensing-controlled behaviors both in mono- and multi-species communities.
Part III: Frontiers - Quorum Sensing and Disease
-
Scientists are developing synthetic compounds that disrupt bacterial quorum sensing by blocking autoinducer production or detection.
-
Disabling quorum sensing makes bacteria unable to act as collectives, which decreases bacterial pathogenicity, biofilm formation, etc.
-
Quorum-sensing interference strategies are also found in nature. In mixed bacterial communities, chemical “warfare” is occurring, which allows particular bacteria to cheat, free ride, eavesdrop, and send misinformation!
-
These natural quorum-sensing interference strategies (called quorum quenching) are being used as inspiration to develop antimicrobial medicines, surface coatings that prevent bacterial biofilm formation, and other products for health, industry, agriculture, and the environment.
-
Another horizon for scientists is to discover how quorum sensing works in natural, rather than laboratory, contexts. Challenges include learning how quorum sensing operates in heterogeneous environments that fluctuate in time, space, and bacterial species composition as well as contain eukaryotic hosts and viruses.
Closing Thoughts
-
No one could have imagined initially that the study of bioluminescence in bacteria would lead to new ideas for biotechnology. Curiosity-driven research often lays the groundwork for pragmatic applications.
Guided Paper
Papenfort, K., Silpe, J.E., Schramma, K.R., Cong, J.P., Seyedsayamdost, M.R., and Bassler, B.L. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nature Chem. Biol. 2017;13:551–557.
This paper describes the discovery of a new quorum-sensing mechanism (and a new autoinducer called DPO) in the pathogenic bacterium Vibrio cholerae, as discussed in the Frontiers section of this Narrative.
DownloadIntroduction
Bacteria (also known as prokaryotes) are Earth’s most ancient organisms. They are most famously known as scourges that cause deadly diseases. However, what has been discovered recently is that bacteria are also life-giving. Large communities of bacteria live in and on higher organisms (such as you) in assemblies called microbiomes. Microbiomes are crucial for human, animal, and plant health because microbiome bacteria supply their larger hosts with vital substances. Scientists now think that multicellular life on Earth co-evolved with bacteria and that the lives of humans and bacteria are intimately intertwined.
But how can bacteria, which are 500 times smaller than a human cell, kill us, or conversely, give us life? I have spent my life wondering how these tiny critters got the ability to control the fates of outrageously bigger organisms. What scientists found is that bacteria have power in numbers and that they have sophisticated communication abilities that, earlier, were thought impossible for such small, “primitive” organisms. In this Narrative, I will try to convince you that bacteria can “talk” to each other, that they are multilingual, that they act together in coordinated groups, and that these capabilities give bacteria their awesome power.
Bacteria communicate with one another—not with words but with chemicals. Bacteria release these chemical molecules (called autoinducers) into their environment and then they use the buildup of these signaling molecules to take a census of their cell numbers. When a critical number of cells is reached, the bacteria recognize that they are in a group and they behave as a coordinated team exhibiting new behaviors.
In the time-lapse movie shown here (Video 1), a particular bacterial species (normally ocean-dwelling) is growing on a dish in a laboratory. When the bacterial cells reach a critical number, they start producing blue light, a process called “bioluminescence.” The release of chemical molecules called autoinducers (normally invisible but depicted in the cartoon) causes the bacteria to switch on light production. This process is called “quorum sensing.” In the Journey to Discovery, we will learn how scientists discovered quorum sensing and how they uncovered the autoinducers and their interacting “receptors” that underlie this phenomenon.
Quorum sensing allows bacteria to collectively accomplish tasks that would be unsuccessful if a single bacterium acted alone (Figure 1). In addition to bioluminescence shown in the movie, quorum sensing controls the production of toxins that are crucial for bacteria to cause disease. Quorum sensing controls biofilm formation. Biofilms are communities of bacterial cells adhered to surfaces and a predominant form of bacterial life on Earth. Biofilms help many bacteria to act as pathogens and cause disease. Biofilms are also critical for beneficial bacterial behaviors such as symbiosis as well as in industrial applications, such as waste-water purification and bioremediation. Quorum sensing controls competence, which is the process that enables bacteria to acquire DNA from other cells. Competence allows bacteria to diversify their genomes and has been spectacularly exploited to get bacteria to harbor foreign genes in the biotechnology industry.
Bacteria use multiple quorum-sensing autoinducers for communication in complex environments composed of many bacterial species, which is the normal situation in your intestine, mouth, skin, and other environments. They decode these chemical blends to distinguish self from other and, we think, in so doing, they discern friend from potential foe. This capability allows bacteria to form teams with the correct, helpful, team members, and avoid being duped by enemy bacteria in the neighborhood that might be trying to take advantage of them. Knowing self from other is crucial throughout all of biology.
In the Knowledge Overview, I will discuss how quorum sensing works through a process called signal transduction, which is triggered by autoinducer molecules binding to receptors (specific detector proteins), which subsequently leads to large numbers of genes being turned on or turned off. Only by first deeply understanding how bacteria communicate and the kinds of behaviors that are controlled by quorum sensing, can scientists think up cunning ways to tinker with the process, and in doing so, invent quorum-sensing-manipulation applications that will be successful. In the Frontiers section, I will provide some examples of the ways that scientists are using their understanding of quorum sensing to improve agriculture and fight disease.
I hope you will read on. My dream is that this text will make you feel as if you are looking over my shoulder, over the shoulders of three important scientists who came before me, and over the shoulders of the newest generation of scientists as we go on adventures. I hope you will understand and feel how we felt as we did our experiments, interpreted (and misinterpreted) our results, went down wrong paths, and also went down successful paths, as we made our discoveries showing that bacteria can “talk” to each other.
Part I: Journey to Discovery —
Quorum Sensing and the Molecules Involved
The Problem
Bacteria are the world’s most intensively researched organisms, having been studied for nearly 400 years. Bacteria were the model organisms used for famous studies showing that DNA is the molecule conferring heredity (see Whiteboard Video on the Avery, MacCleod, McCarty experiment) and that spontaneous mutations are the basis of evolutionary selection (see Narrative on Mutations by Koshland).
Until about 25 years ago, however, bacteria were considered to act strictly as individuals in that they were incapable of cell-cell communication and group behaviors. The long-standing dogma was that bacteria were too primitive to be able to accomplish feats such as collective social behaviors. Such “fancy traits” were generally considered to be reserved for higher organisms such as bees and man. This thinking probably held back the field.
To me, one of the best things about being a scientist is that dogma can be overturned and scientific fields can change. It might take a while, but good data lasts over time, and scientists are persuaded by convincing data. Scientists, by nature, are skeptics. That is a fantastic character trait because it means scientists think critically about information and models can be overturned by sound, data-driven proof. That is what happened in the quorum-sensing field. Now we know that quorum sensing is the norm, and not an exception, in the bacterial world.
There were a few pioneering scientists who performed compelling experiments that demonstrated that bacteria can communicate with one another. In the Journey to Discovery, I will feature four experiments. Two experiments were done by the first pioneers—Alexander Tomasz and Woody Hastings—their work provided the foundation for the field. The second two experiments involved my “scientific father” Mike Silverman, who was my postdoctoral advisor. The first of these discoveries was made by a graduate student, Joanne Engebrecht, and Mike Silverman in a beautiful experiment that led to the initial molecular mechanism underlying bacterial cell-cell communication. The second discovery was made by myself and Mike Silverman and it led to the discovery that bacteria are “multilingual” possessing vocabularies involving multiple chemicals. As this was my own work, I can also explain how I went down the wrong path before the correct and more exciting path.
Clues
Clue 1: The experiment of Alexander Tomasz shows that bacteria can communicate with one another using small molecules.
Under particular conditions, bacteria can take up DNA from their surroundings, a process called “transformation” (Figure 2) discovered in 1944 by Avery-MacLeod-McCarty (see Avery, Macleod, McCarty Whiteboard). DNA transformation allows bacteria to acquire and propagate genes, which formed the basis of much of modern DNA cloning technology.
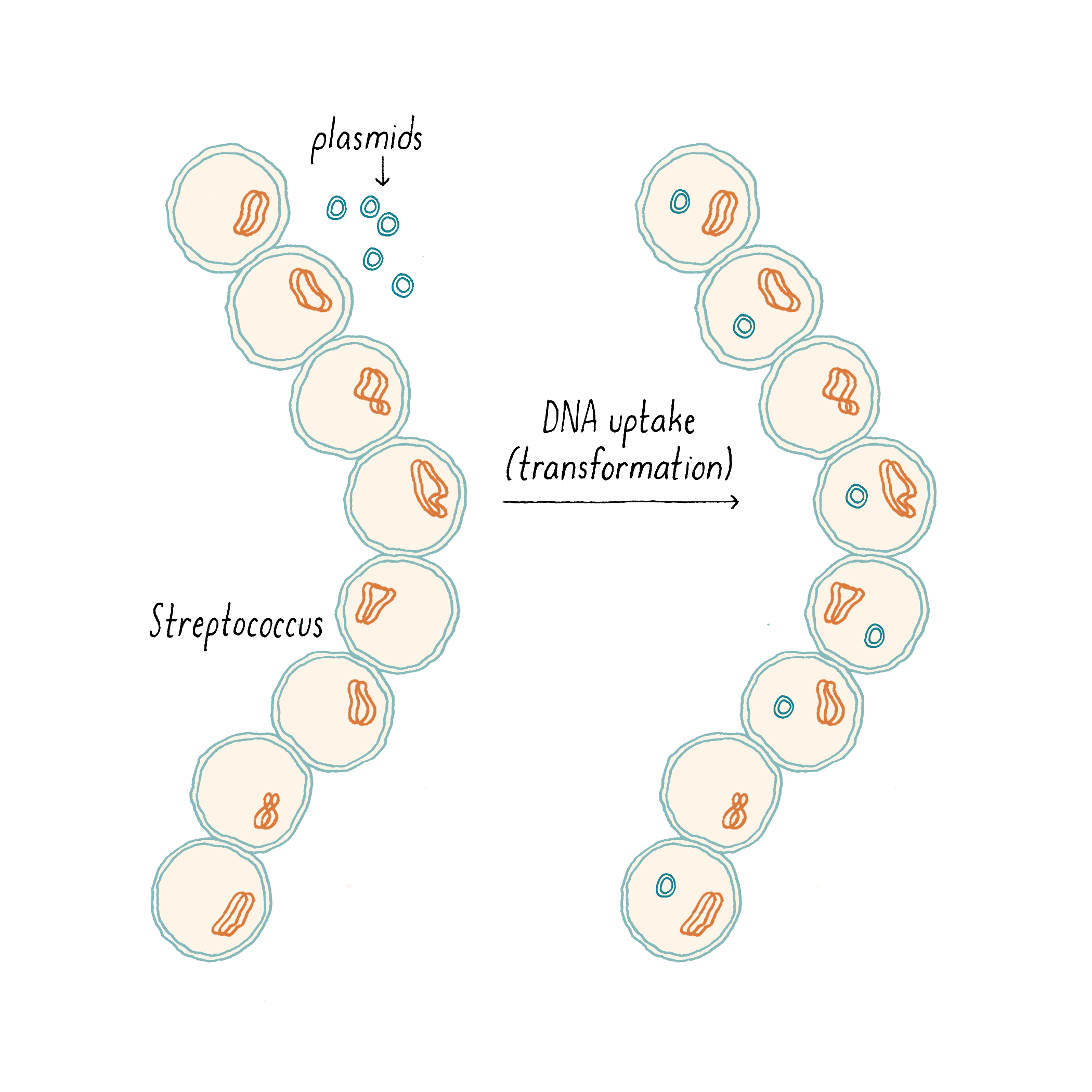
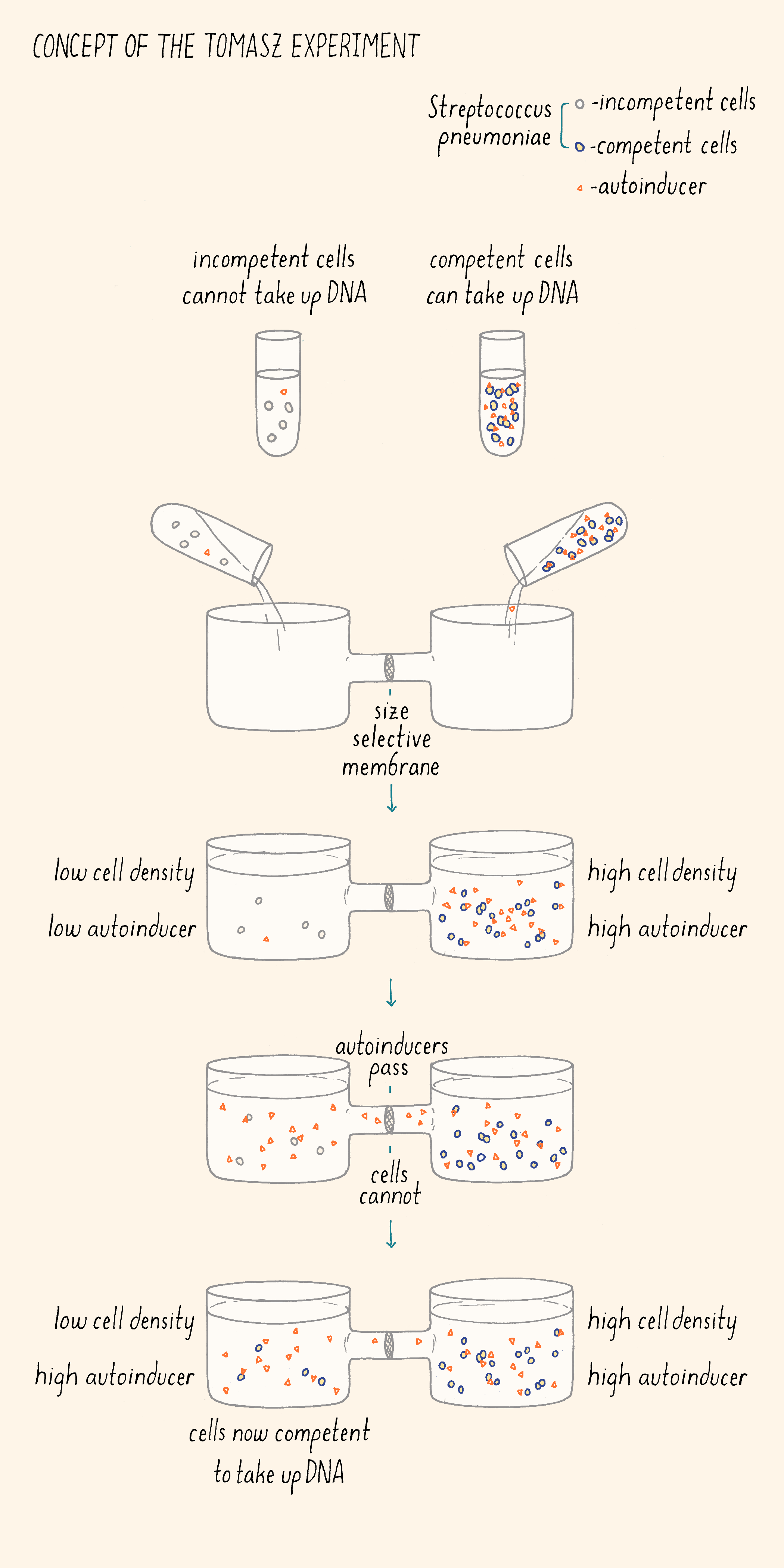
In 1965, Alexander Tomasz was studying how the bacterium Streptococcus pneumoniae took up DNA from the environment. It was known that S. pneumoniae needed to enter a so-called “competent state” in order to acquire DNA. It was also known that, in growing S. pneumoniae cultures, most of the cells became competent to take up DNA during only a short window of time, at a particular phase in the growth curve, and, most importantly, the cells became competent synchronously. To explore how the cells managed this, Tomasz did a number of experiments, here are two of them.
First (Figure 3), he collected competent and incompetent S. pneumoniae cells. He put the two kinds of cells in two chambers containing growth medium separated by a filter. The filter allowed small molecules to pass between the two chambers but the competent and incompetent cells could not come in direct contact with one another. He showed that the incompetent cells became competent when located in the chamber adjacent to the competent cells, suggesting something (a molecule) was passing through the filter from the competent cells to the incompetent cells. In his next experiment, Tomasz isolated the growth medium from competent cells, concentrated the components in it, and added that preparation to incompetent cells. The incompetent cells became competent! These experiments demonstrated that the ability to become competent was regulated by an extracellular factor that was produced by Streptococcus itself. He described this competence factor as a “hormone-like activator” that synchronizes the behavior of the bacterial population.
Tomasz wrote,
“Since the activator—a cell-produced chemical—seems to impose a high degree of physiological homogeneity in a pneumococcal population with respect to competence, one is forced to conclude that in this case [a] bacterial population can behave as a biological unit with considerable… coordination among its members.…”
He wrote another amazing sentence in this manuscript, almost 30 years before the quorum-sensing field came into vogue:
“One wonders whether this kind of control may not be operative in some other microbial phenomena also.”
Below, I will provide some thoughts about why this finding and this central idea did not immediately inspire the imaginations of other scientists.
Clue 2: Hastings and co-workers uncover evidence that an “autoinducer” controls bioluminescence.
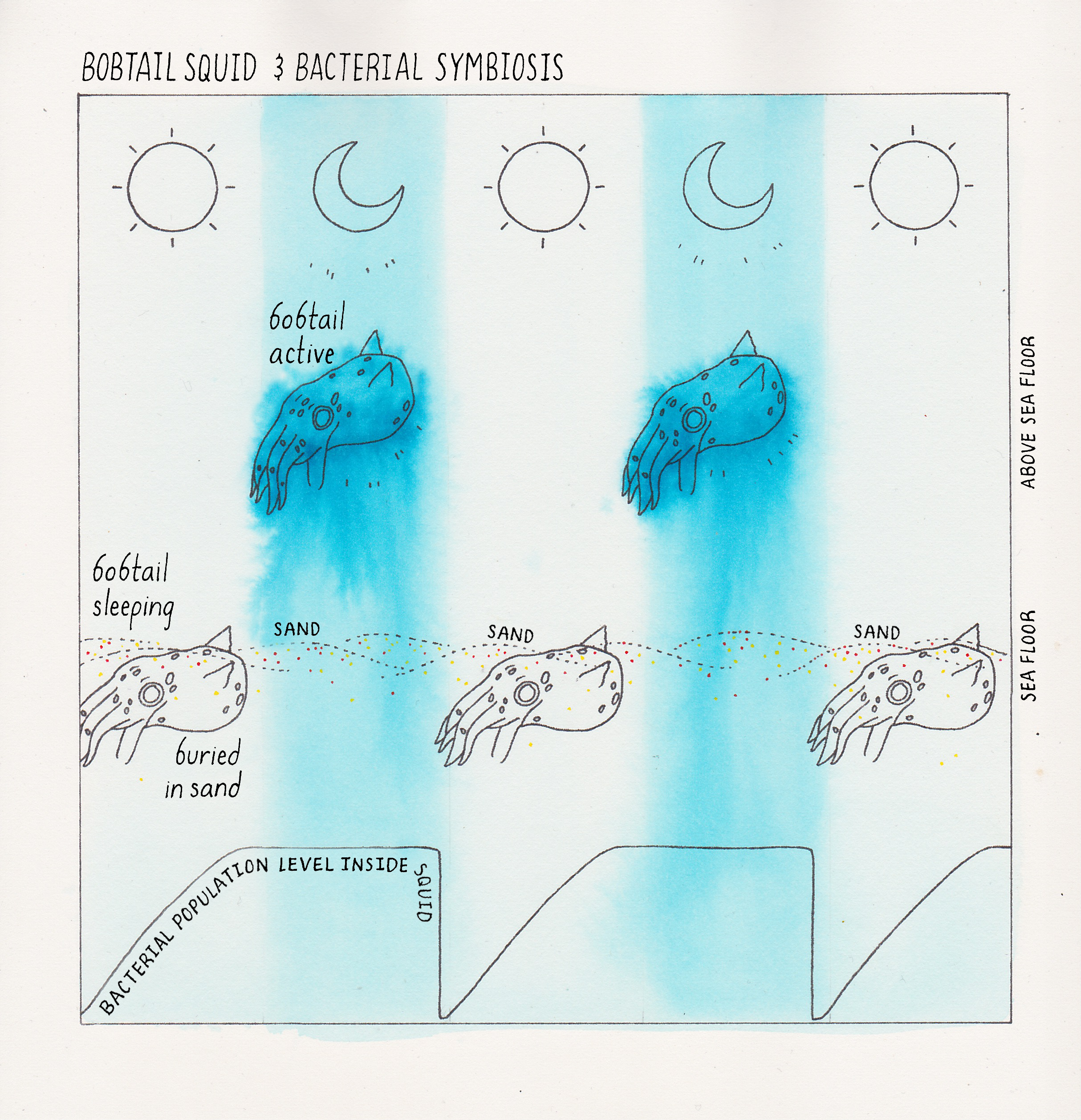
Perhaps unaware initially of the earlier work on DNA competence, Woody Hastings and his coworkers performed experiments that independently led to similar conclusions as those of Tomasz. Hastings was fascinated by all things bioluminescent —marine bacteria, dinoflagellates, fungi, fireflies, fish, etc. He wanted to understand how these organisms could make light. In the example that is relevant here, Hastings was studying an obscure marine bacterial species, Vibrio fischeri. V. fischeri exists in many habitats in the ocean, among them as a symbiont of marine animals. In these symbioses, the bacteria provide light that the animal host uses for some purpose, such as attracting a mate, scaring away predators, attracting prey, and so on. In turn, the animal provides V. fischeri more nutrients than are typically found in the free ocean. Because both partners- V. fischeri and the animal—benefit, we call this relationship a symbiosis.
We highlight one example here (Figure 4). V. fischeri can form a symbiotic partnership with the Hawaiian Bobtail squid (Euprymna scolopes). E. scolopes is a nocturnal animal that lives in shallow (knee-deep water). The squid buries itself in the ocean floor during the day to stay hidden from predators (Figure 4). At night, E. scolopes emerges from the sediments to hunt for food in the shallow, knee-deep water, but it is now also vulnerable to predation. That is because, on clear nights, when the moon is out, the moonlight shining down onto the squid casts a shadow on the seafloor a few feet beneath the squid. Predators could see the shadow and be alerted to the squid’s presence. This is where V. fischeri comes in. The light made by the V. fischeri cells housed in the squid’s light organ is cast onto the seafloor below the squid. Shining the light downward obscures any shadow the squid might make due to the moonlight, effectively making the squid “invisible” to its enemies. In this amazing symbiotic relationship, V. fischeri provides the squid with bioluminescence to help make the squid less visible to predators. On the other side of the symbiosis, the squid light organ is loaded with nutrients that allow V. fischeri cells to flourish and grow to vastly higher numbers than are achievable in the open ocean which is a nutrient desert. (Want to know more about this symbiotic relationship? See Dig Deeper 1 and the iBiology talk by Margaret McFall-Ngai).
In 1970, Hastings and his mentees Ken Nealson and Terry Platt were growing V. fischeri in the laboratory to study its bioluminescence. They did a simple experiment in which they measured light production over time and how many cells were present at each time point during the growth of the V. fischeri bacteria in the culture (Figure 5). What they found was that the bacteria produced bioluminescence only at high cell density, but not when the V. fischeri cells existed in dilute (low cell density) suspensions. You can hear Dr. Hastings discuss this experiment in his talk on iBiology.
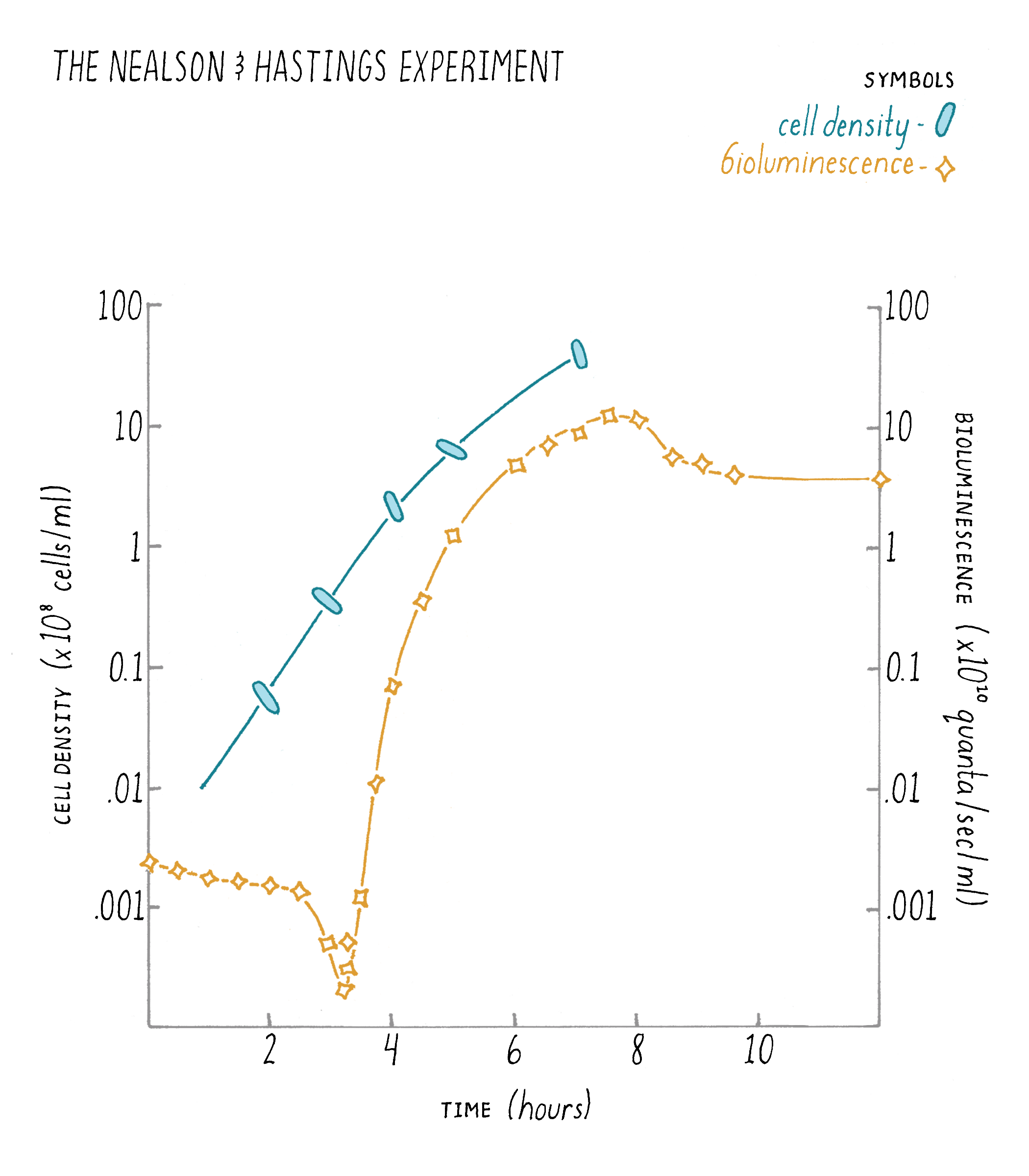
Explorer’s Question: In the graph, what do you notice about the differences in the cell growth rate and the corresponding increase in light produced by the cells?
Answer: The main point to notice is that bioluminescence increased faster than could be accounted for by cell doublings. That is, the cells would double in number (2-fold) but in the same period of time, light would increase 10-fold. This curious discrepancy led the researchers to suggest that a cell-produced inducer must be released into the culture medium, that it accumulated, and was involved in accelerating light production by the cells (Nealson et al., 1970). Nealson called this substance an “autoinducer".
An astute explorer also might notice a dip in the bioluminescence at early time points. When the scientists diluted the high cell density V. fischeri cells to start the experiment, they also diluted the autoinducer that had accumulated outside of the V. fischeri cells during the previous growth period. This dilution step reduced the concentration of extracellular autoinducer to below the level required for the cells to detect it. V. fischeri responds to this sudden loss of autoinducer by shutting off light production. For the first few hours of the experiment, light production declines because no new luciferase enzymes (light-producing enzymes) are made. Each time a cell divides, half of the existing luciferase enzymes end up in one cell and half ends up in the other cell. Thus, the total concentration of luciferase per cell decreases by half every time the cells double. Hence, the dip in light production in the curve in the figure. However, the V. fischeri cells are growing during this time, and they are making and releasing new autoinducer into the medium. That newly made autoinducer accumulates, and when it once again reaches the level required for detection (after about 3 hours of growth in the figure), the V. fischeri cells commence making light, which, as you see in the graph, shoots up rapidly.
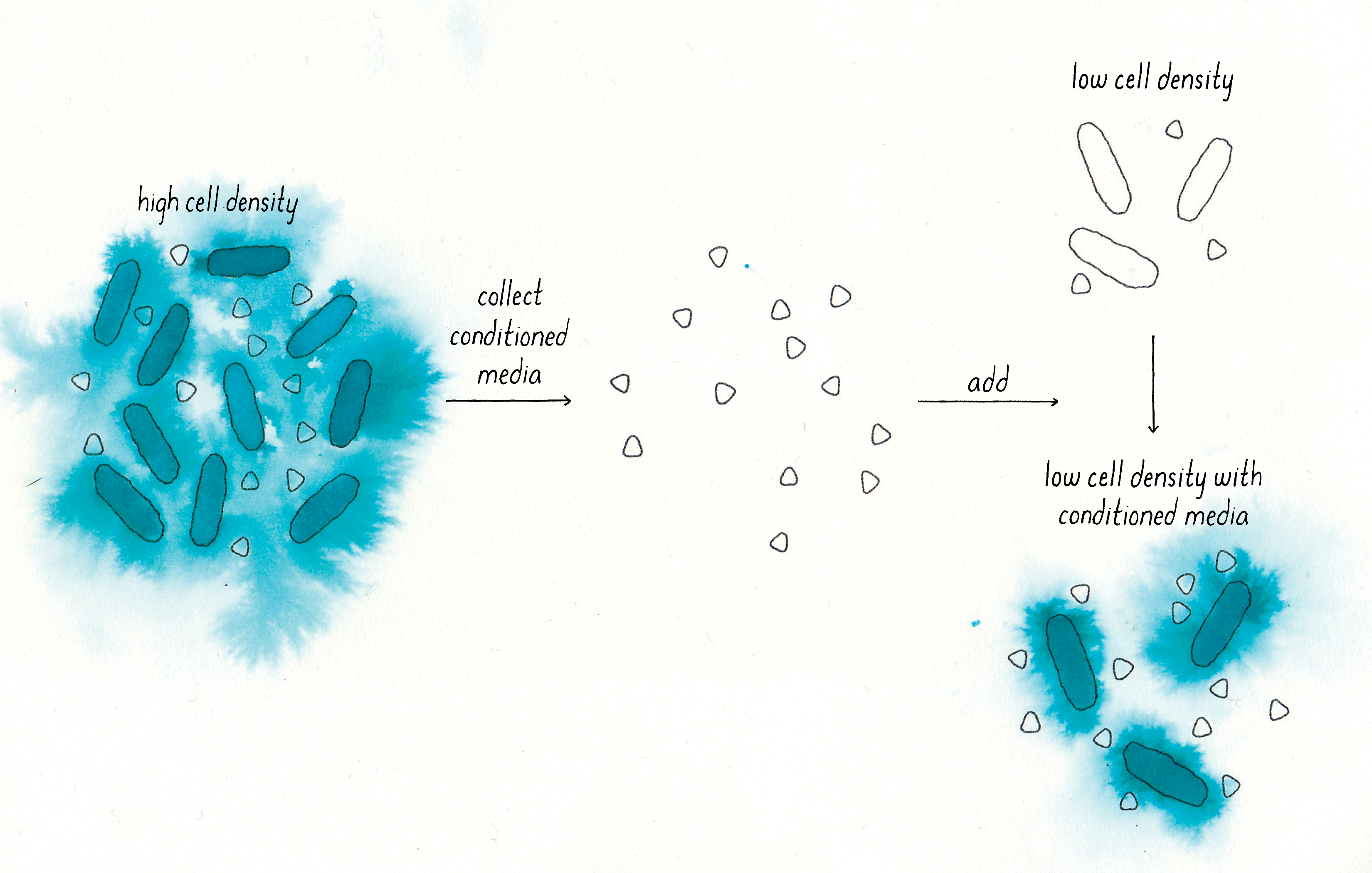
Explorer’s Question: What would you do next? How might you confirm the hypothesis that high cell density V. fischeri cells secrete an “autoinducer” molecule?
Answer: Nealson showed that the autoinducer existed in a later manuscript when he did a clever experiment—he effectively “tricked” low cell density V. fischeri cells (no light production) into behaving like high cell density V. fischeri cells (producing bioluminescence). He did this by collecting “conditioned” medium from high cell density V. fischeri cells (removing the cells and leaving only the solution that the cells had been grown in) and then adding it to the low cell density cells (Figure 6). When he did this, the low-density cells began to glow! This experiment showed there must have been a stimulatory molecule (an autoinducer) present in the high-cell density culture fluids. This finding is similar to that from the Tomasz experiment that used conditioned medium.
Eberhard and Nealson, both of whom trained with Hastings, then began the hunt for the mysterious autoinducer substance. Eberhard was a terrific chemist. He fractionated V. fischeri high-density conditioned media preparations and tested each fraction for the ability to induce light production in low-density V. fischeri cells. Tracking the activity by this method allowed him to isolate the compound, and, using companion chemical analyses, Eberhard determined its structure. In this first case, he identified the molecule as a compound called a homoserine lactone. We will come back to the structure of the autoinducer later.
The Discovery: Quorum Sensing in Bacteria
Discovery A: Finding genes involved in quorum sensing
The identification of the autoinducer was important, but how did it work? That question was tackled by Mike Silverman and his graduate student Joanne Engebrecht in 1983. Silverman knew the V. fischeri findings, and he reasoned that light production could be used as a powerful built-in, visible marker of the bacteria communicating with the autoinducer.
Engebrecht and Silverman designed a cunning but fantastically simple experiment to identify the components that enabled V. fischeri to glow (Figure 7). Their strategy: take DNA from V. fischeri and transfer pieces of it into E. coli (a bacterium that normally lives in your gut, and which does not produce bioluminescence) and then check for glowing E. coli, presumably containing a piece of V. fischeri DNA with important genes required for light production. Of course, they did not know which V. fischeri genes were responsible for bioluminescence itself, for making the autoinducer, or for the response to the autoinducer. That was the point of the experiment. So, they sheared the V. fischeri genome into fragments and cloned all of the segments into plasmids. They introduced this “library” of plasmids (each plasmid containing a random fragment of V. fischeri genomic DNA) into E. coli. They grew the E. coli into colonies on Petri plates, turned the lights off in the room, and in the darkness, they scanned their Petri plates for a recombinant E. coli colony that glowed in the dark (i.e., that emitted bioluminescence).
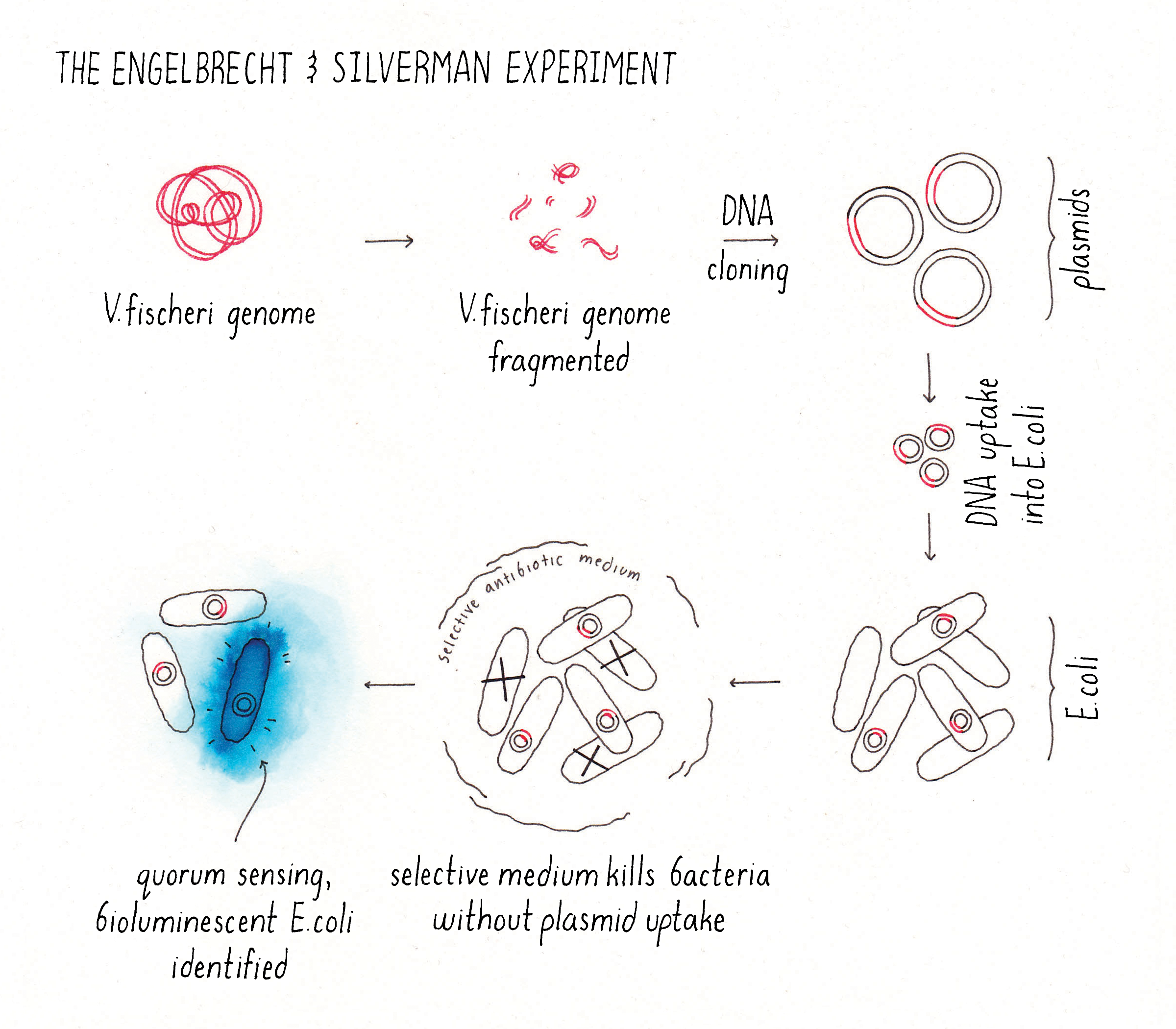
Explorer’s Question: What problem(s) might have Engebrecht and Silverman encountered in their experiment?
Answer: The biggest unknown was how many genes were required for making autoinducer, the receptor, and bioluminescence (light production) itself. And they did not know if the genes could work in isolation or if all the genes needed to function together to enable E. coli to make light. What they did know is that bacteria often harbor groups of genes with related functions close to one another in the genome (these clusters are called operons). They rationalized that if the autoinducer production and response genes and the light-making genes were co-located on the DNA, they would all be transferred into E. coli as a unit and their strategy could work. But if there were many genes involved, the likelihood that they would all be adjacent to one another on the same genomic DNA fragment for transfer into E. coli would be low. Engebrecht and Silverman took the gamble and tried the experiment, and it worked!
Remarkably, Engebrecht and Silverman found an E. coli colony that glowed in the dark (Figure 7)! They isolated the fragment of V. fischeri DNA from the glowing E. coli colony and sequenced it to identify the genes contained on the DNA fragment. This analysis showed that the DNA fragment contained several genes encoding the following proteins: (1) the enzyme that makes the autoinducer, (2) an autoinducer receptor (the protein that binds and detects the autoinducer), and (3) the proteins that make light (an enzyme complex called luciferase). Consistent with this analysis, they showed that their bioluminescent E. coli produced the autoinducer and released it into the culture fluids and their E. coli only emitted bioluminescence at high cell density. Thus, in one elegant experiment, and on one single fragment of DNA, Engebrecht and Silverman identified the genes for cell-cell communication, and the genes for light production and the entire system functioned properly in E. coli. Amazing!!
Silverman named the genes required for light production “Lux,” for Luminescence expression with the idea that people would be reminded of Luxor the Egyptian city housing the temple for the God of Light. Engebrecht and Silverman came up with “LuxI” as the name for the “Inducer” (more accurately LuxI is the enzyme that synthesizes the autoinducer), and “LuxR” for the “Receptor” that binds the autoinducer. Engebrecht and Silverman proposed a brilliant model (Figure 8), which is discussed below:
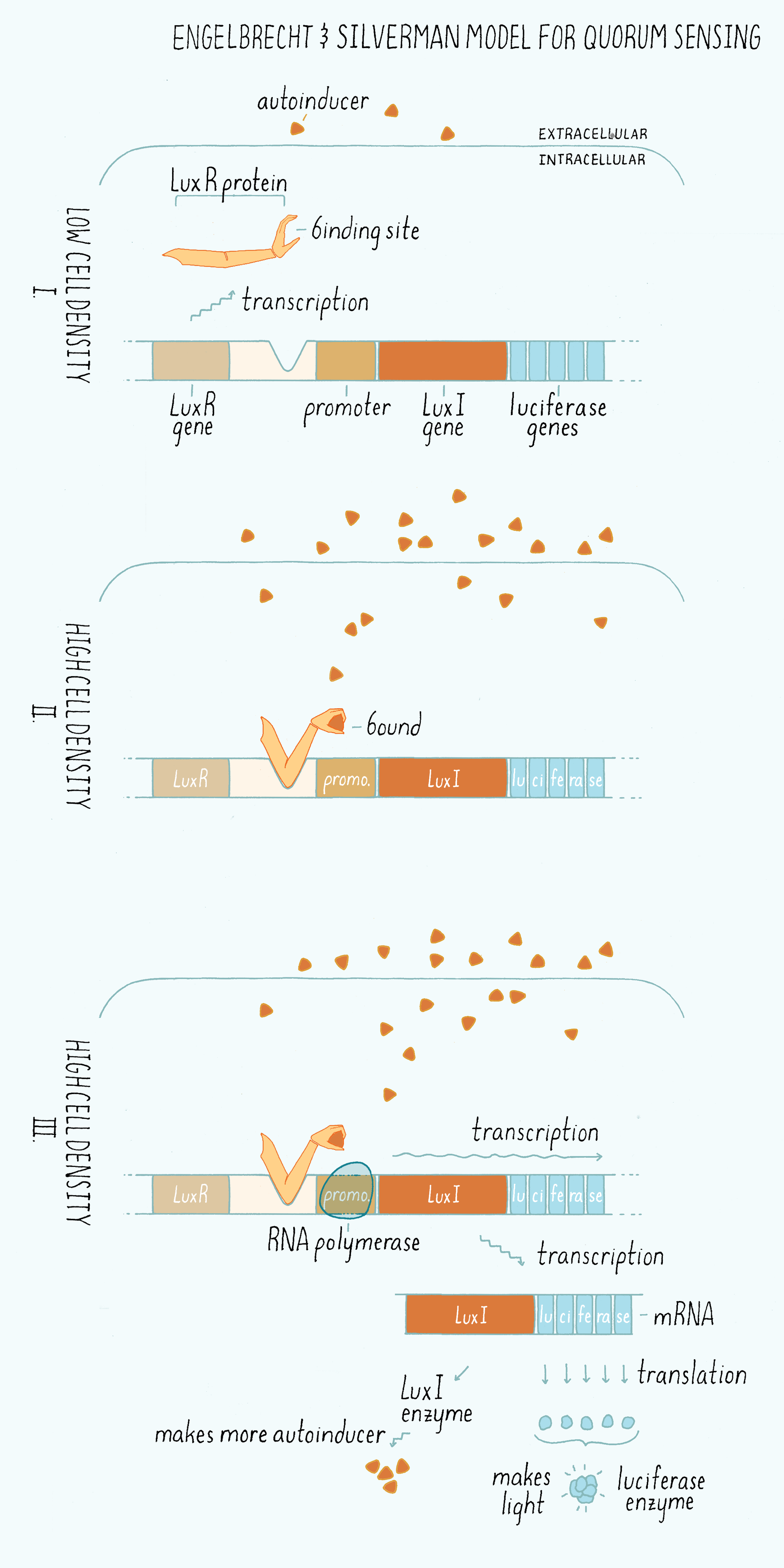
1) The enzyme (called LuxI) makes the homoserine lactone autoinducer. The homoserine lactone is a small molecule that can freely cross the plasma membrane and thus pass into and out of the cells. The autoinducer concentration increases in parallel with increasing cell density.
2) Only at high cell density, is sufficient autoinducer present for it to be bound by the receptor (LuxR) in the cytoplasm.
3) LuxR is an activator of transcription. Only when LuxR is bound to the autoinducer molecule can the LuxR-autoinducer complex bind to a region of the DNA called a promoter and activate the transcription of the genes encoding luciferase, the light-producing enzyme complex. When luciferase is made, the cells make light.
4) The gene encoding the enzyme that makes the autoinducer (LuxI) is also located in the same operon as the luciferase genes. So, when the receptor-autoinducer complex is bound to DNA, additional LuxI enzyme is made and an additional autoinducer molecule is likewise made. This increased autoinducer binds to the LuxR receptor, which then binds DNA further increasing luciferase gene expression. This feature “amplifies” and “accelerates” the output (light production). This genetic arrangement, in which a component turns on more of its own synthesis is called a “positive feedback loop,” which we will discuss more in the Knowledge Overview.
Explorer’s Question: Now that you know how the V. fischeri system functions and you know about how autoinducer accumulation leads to more autoinducer production and, in turn, increased response to the autoinducer, does that help you to better understand the data in Hasting’s original experiment (Figure 5)?
Answer: What you see in the graph is that every time the cells double in number, the light turns on about 10-fold (not 2-fold as you might have predicted). This effect occurs because of the positive feedback “autoinduction” loop. Cells that commence the quorum-sensing behavior of producing light, also greatly boost their autoinducer production levels. This action “floods” the vicinity with a high-level autoinducer causing all the nearby cells to rapidly turn on light production. They too turn on increased autoinducer production further amplifying the effect. The ultimate result is that light production increases much faster than can be accounted for by each cell dividing and merely doubling the amount of luciferase made. We see this asymmetry over and over in biology and it is a hallmark of positive feedback loops. We will discuss these feedback loops in the Knowledge Overview.
Together, Tomasz’, Hastings’, and Silverman’s initial discoveries, which spanned 1965–1983, suggested that at least two species of bacteria used the production, release, exchange, and detection of small molecules (autoinducers) to measure their cell numbers and to control group behaviors in response to changes in cell population density.
For another decade, these cell-to-cell communication discoveries were considered anomalies. Likely, the clues were not well connected. Tomasz’ experiment, which was much earlier and with a very different bacterium, was not thought to be related to Hastings’ and Silverman’s findings with bioluminescent bacteria. Indeed, a full 30 years would pass between Tomasz’ discovery and the identification of the S. pneumoniae autoinducer molecule. With respect to the Hastings and Silverman findings, V. fischeri was neither a commonly used bacterium in research nor thought to be of medical, industrial, or environmental relevance. The research was not actively picked up by others, because the findings were not considered “highly “relevant.”
Discovery B: Bacteria are multi-lingual
The second discovery story of this Narrative is my own, which I also describe in Video 2.
The common character in these two stories is Mike Silverman, my postdoc advisor from 1990 to 1994, who is featured in the discovery story above. (see Dig Deeper 2 to learn about how I became Silverman’s postdoc).
Silverman and I wanted to find out if there were other, interesting components involved in quorum sensing that were not discovered in his original experiment. As you understand from Silverman and Engebrecht’s experiment, they had transferred V. fischeri genes into E. coli and looked for bacteria that began to glow. We wanted to do the opposite: take bacteria that normally glowed in response to autoinducers and introduce mutations that would cause them to stop glowing (for more information, see the Narrative on Mutations by Koshland. We reasoned that we might be able to find additional genes by this method.
Alas, we did not have tools to genetically manipulate V. fischeri, which was essential for our strategy. So we had to choose a new model organism. Silverman and I decided to work on Vibrio harveyi, a bioluminescent marine bacterium that is closely related to V. fischeri. V. harveyi exists free-living in the ocean and apparently has no symbiotic hosts. However, we knew that V. harveyi, like V. fischeri, only produced light at high cell density, and we knew that V. harveyi produced an autoinducer and released it into the cell-free conditioned medium. This was crucial in our decision making; Silverman had gotten genetic technologies working in V. harveyi, meaning that we could transfer DNA into V. harveyi and have it become incorporated in the genome.
So off we went in search of mutations that disrupted bioluminescence in V. harveyi. My strategy was to use transposon mutagenesis (Figure 9). A transposon is a small DNA sequence that can insert randomly into the bacterial genome. If the transposon lands in a gene, then that gene is mutated and it can no longer make a protein. I made tens of thousands of mutant bacteria each with a unique transposon insertion somewhere in the genome. I thought that I should scan all the mutant V. harveyi colonies for those that made no light because those bacteria would have obtained a transposon insertion in a gene required for light production or for the regulation of light production. The plan was to isolate those non-light-producing bacterial colonies and then identify the genes in which the transposons had landed.
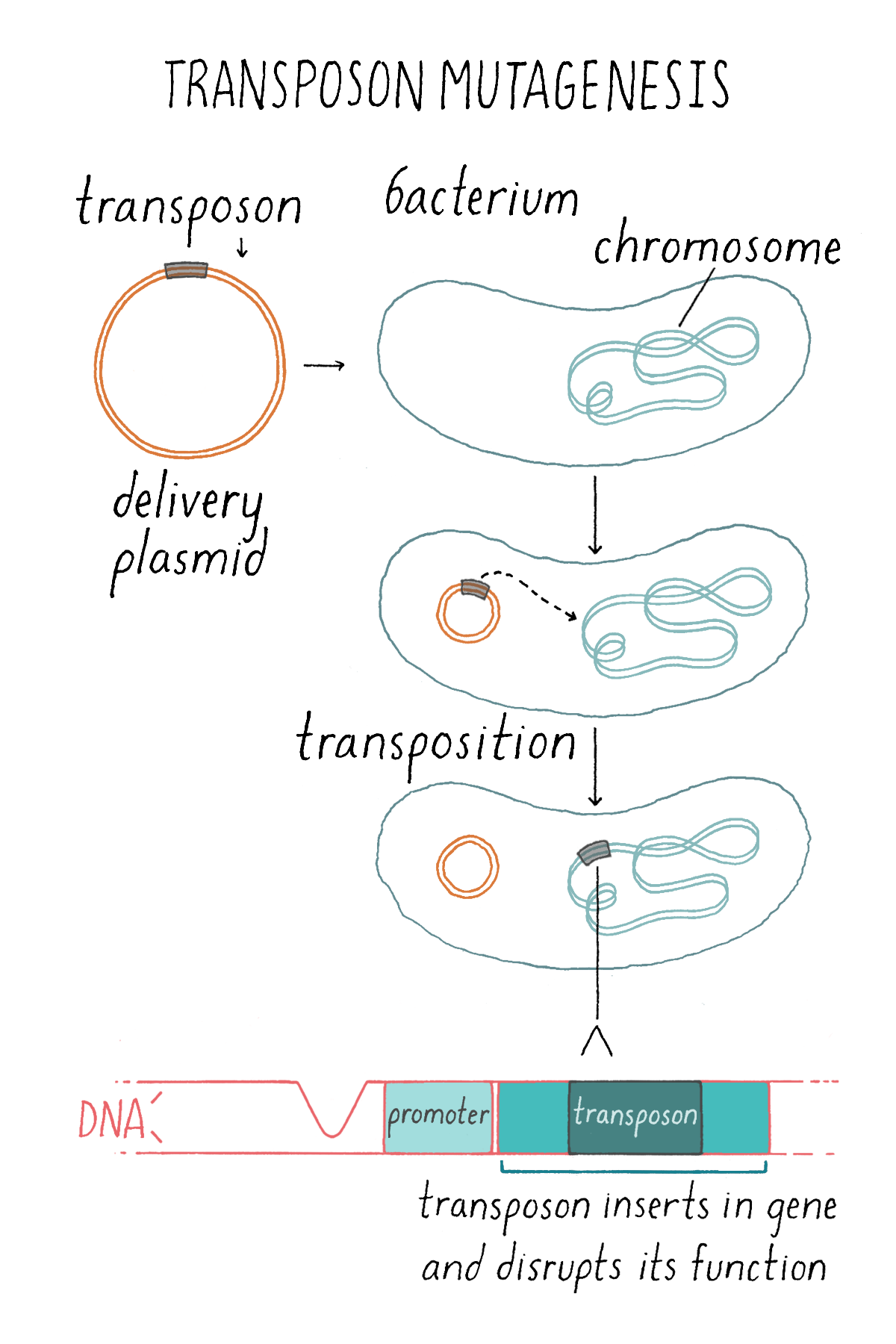
The explorer (myself) made one overarching assumption, which turned out to be COMPLETELY WRONG, but as you will see, it had a happy ending. I thought that as V. harveyi and V. fischeri were so similar—they both made and responded to an autoinducer to control light production in response to increased cell density—they would both possess luxI (autoinducer production gene), luxR (autoinducer receptor gene), and luciferase genes. And, I reasoned, therefore, that each of those genes would be absolutely required for light production. My logic: if a transposon landed in any of those genes, then that V. harveyi mutant would be dark (make no light). I was especially interested in identifying luxI and luxR, thinking that, after I found and characterized those known genes, I could dispense with them and then move on to find out if other interesting components were involved.
I made transposon insertions and collected non-light-producing mutants for a year. My characterizations, over and over, showed that all the genes that had transposon insertions were genes encoding luciferase but never, ever in any other gene, such as luxI or luxR, which I expected would regulate luciferase production.
My results made no sense—I knew there was an autoinducer so there had to be a gene for an enzyme that made it. I knew the autoinducer was detected only at high cell density so there had to be a gene encoding a receptor protein. But I did the same experiment over and over and over. Finally, one day, after I again got the same results, I stopped just doing the experiment that was obviously not working, and sat down to think- why I might not be getting mutations in regulatory genes using this strategy?
Explorer’s Question: What possible explanations can you think of for why the experiment did not work?
.Answer: I came up with two ideas. First, if the genes I was after were essential for bacterial life, then, if such a gene got inactivated by the transposon, the bacteria would die and I would not isolate them on my petri plates. That did not make too much sense to me because I could inactive luciferase genes and the bacteria were healthy. So, I reasoned if they did not “need” luciferase, they probably did not “need” the regulatory genes either. A second possibility was that there were two, redundant, regulatory systems. That is, I wondered what if there were two autoinducers and two receptors. Then, in my experiment, if I inactivated a gene for one autoinducer or for one receptor, the other autoinducer-receptor pair could compensate and the bacteria would still make light at high cell density.
Aha! After the “aha moment” finally came, I instantly changed my strategy (that is after I sulked around thinking how could it have taken me so long to come up with this obvious possibility). I decided to look for mutants that were dim but not dark (non-light producing). Dim mutants were obviously capable of making some light, so those mutants could not be defective in luciferase genes (luciferase genes are absolutely required for light). However, as such mutants did not make the correct amount of light, I thought it likely that they might be defective in the proper regulation of light production. I had noticed earlier that dim V. harveyi mutants arose spontaneously at a fairly reasonable frequency. Indeed, in my earlier experiments, I had gotten (and thrown out!) lots of dim mutants that I noticed on my Petri plates. I did the screen again, but this time, I collected the dim mutants from my many tens of thousands of V. harveyi colonies. When I analyzed them, to my delight, I found that V. harveyi had genes encoding not one but two enzymes that make two autoinducers. And I also found genes that encoded not one but two receptors for the autoinducers. However, unlike the LuxR receptor in V. fischeri (Figure 5), these receptors resided at the plasma membrane (Figure 10).
Apparently, V. harveyi had several communication systems functioning in parallel. If one system was gone, the other one did a pretty good job of appropriately regulating bioluminescence in the laboratory. Later, after I started my own lab, I also found a third quorum-sensing system (i.e., another autoinducer production enzyme and a receptor protein pair) in V. harveyi.
In summary, my results showed that bacteria could communicate with multiple autoinducers. That was a huge surprise because it meant that not only could bacteria communicate, but they could do so using several chemical “words” (Figure 10). A second and equally big surprise to us was that V. harveyi used a different signal relay system than V. fischeri. In V. harveyi, the receptors were in the plasma membrane and not in the cytoplasm, as Engebrecht and Silverman found originally for V. fischeri. I will discuss the V. harveyi system and how it works in the Knowledge Overview.
Explorer’s Question: V. harveyi is a free-living marine bacterium, and as I just told you, it uses not one but several autoinducers for communication and control of group behaviors. How can communication mediated by small, diffusible signal molecules ever work in the ocean? The ocean is huge; how can V. harveyi possibly encounter a high enough autoinducer concentration for a response to occur?
Answer: This is my best guess: There are two points you might like to know to help you think about this issue. First, luciferase is not the only trait controlled by quorum sensing in V. harveyi. Indeed, V. harveyi controls about 600 genes using quorum sensing. So, many of its behaviors, in addition to bioluminescence, are carried out only when the bacteria are acting as a group. Second, V. harveyi can exist free swimming in the ocean, but it also frequently lives in structures called biofilms, which are communities of bacterial cells adhered to surfaces. In the ocean, surfaces include sediments, marine snow, marine animals, etc. Biofilm cells are embedded in a matrix, a sort of suit of armor but made up of biological components. The matrix keeps the cells together, keeps them stuck tightly to the surface, sequesters small molecules and metabolites so they do not escape/diffuse away, and enables the community to resist perturbations, such as flow.
When a V. harveyi cell is swimming freely in the gigantic ocean, all of the autoinducer molecules that cell produces and releases immediately diffuse away, and so that cell will not be able to detect any autoinducer. However, that is okay because a free-swimming V. harveyi cell is necessarily by itself, that is, at low cell density, and it should not engage in quorum-sensing-controlled group behaviors. However, when V. harveyi cells grow in biofilms, they necessarily coexist with other cells. In biofilms, V. harveyi can grow to high cell densities. Under this condition, many nearby cells are contributing to overall autoinducer production. Presumably, the protective biofilm matrix helps keep the autoinducer molecules near the cells, enabling the autoinducer to accumulate to the level required for detection. Under this condition, quorum sensing is relevant and the bacteria turn on genes underpinning group behaviors. Recall, many genes turn on under this condition and some of them enable the cells to prosper in the biofilm community lifestyle, for example, by collectively acquiring, breaking down, and sharing nutritious food resources and so on.
What Happened Next?
In addition to the discoveries above made in the two marine bacterial species, three additional laboratories (Williams, Iglewski, and Farrand) discovered quorum sensing in three terrestrial pathogens. In each pathogen, there was an equivalent of LuxI and LuxR that made up autoinducer-receptor pairs. In each case, the autoinducers activated the production of particular proteins at high cell density. However, instead of enzymes for bioluminescence, the induced proteins made the bacteria more virulent (better at causing illness in eukaryotic hosts). Thus, only when the cells were in a group, did they behave as pathogens. Apparently, communication was not simply an oddity of bioluminescence but was relevant for infection.
Together, Engebrecht’s and my work with Silverman, along with the discoveries of autoinducer-receptor pairs in the three terrestrial pathogens mentioned above, proved that (1) cell-cell communication was not restricted to bioluminescent bacteria, rather, cell-cell communication and collective behaviors could be general phenomena in bacteria, (2) markedly different kinds of communication systems had evolved in bacteria, (3) bacteria could detect and make sense of multiple chemical “words,” and (4) cell-cell communication controlled traits that enable certain bacteria to cause disease. The field then called this process “quorum sensing” (See Dig Deeper 3 to learn how the term “quorum sensing” was coined).
In the past 25 years, many quorum-sensing systems have been investigated. Also, with thousands of bacterial genomes sequenced, now scientists use computers to look for DNA sequence matches to known quorum-sensing genes (see Whiteboard video on Bioinformatics). These analyses show that it is very common for bacteria to possess quorum-sensing systems and, moreover, to use multiple autoinducers and multiple receptors for quorum sensing.
Part II: Knowledge Overview —
How bacteria talk to one another
The Basics of Quorum Sensing
Here are the general rules for quorum-sensing systems:
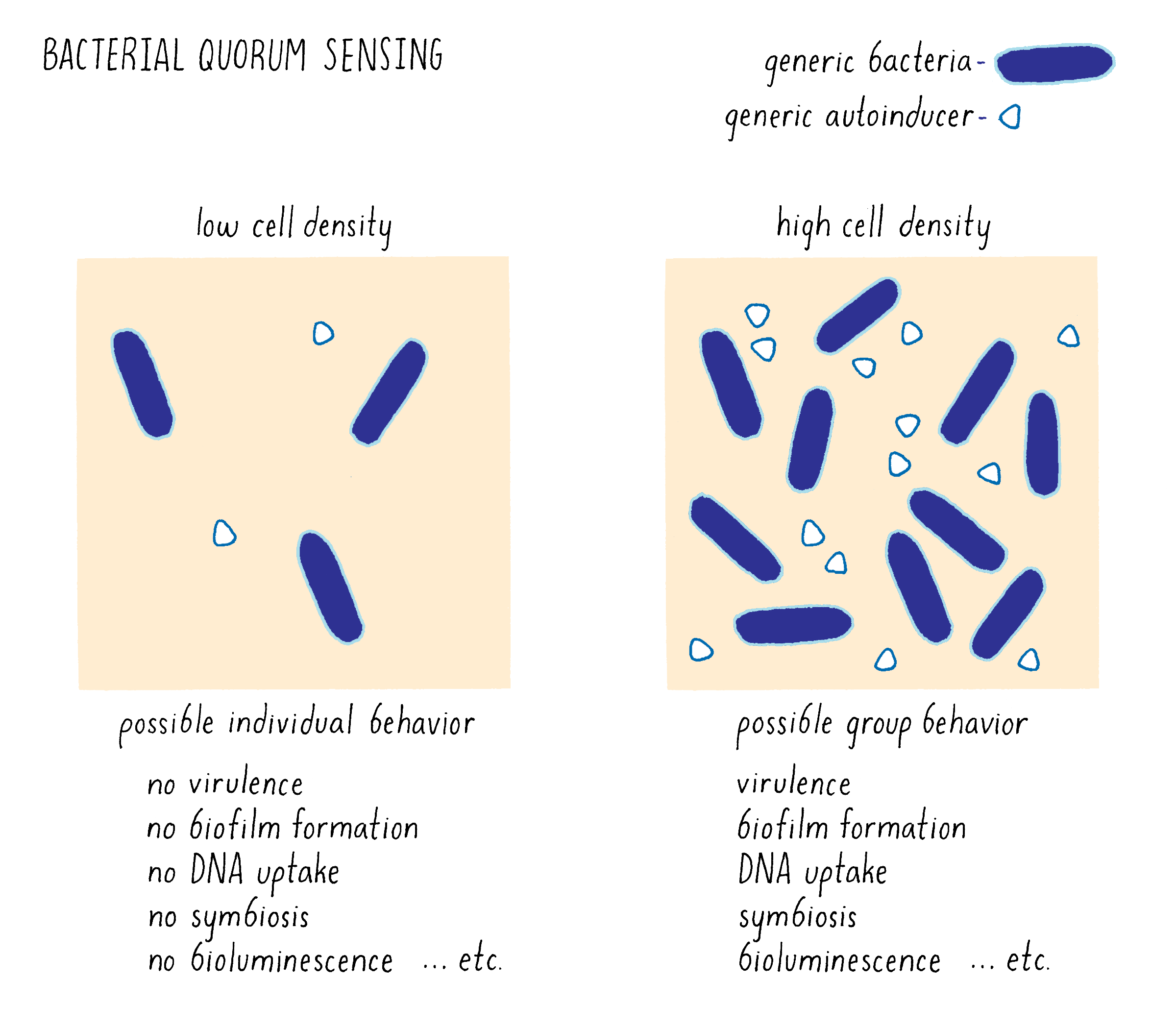
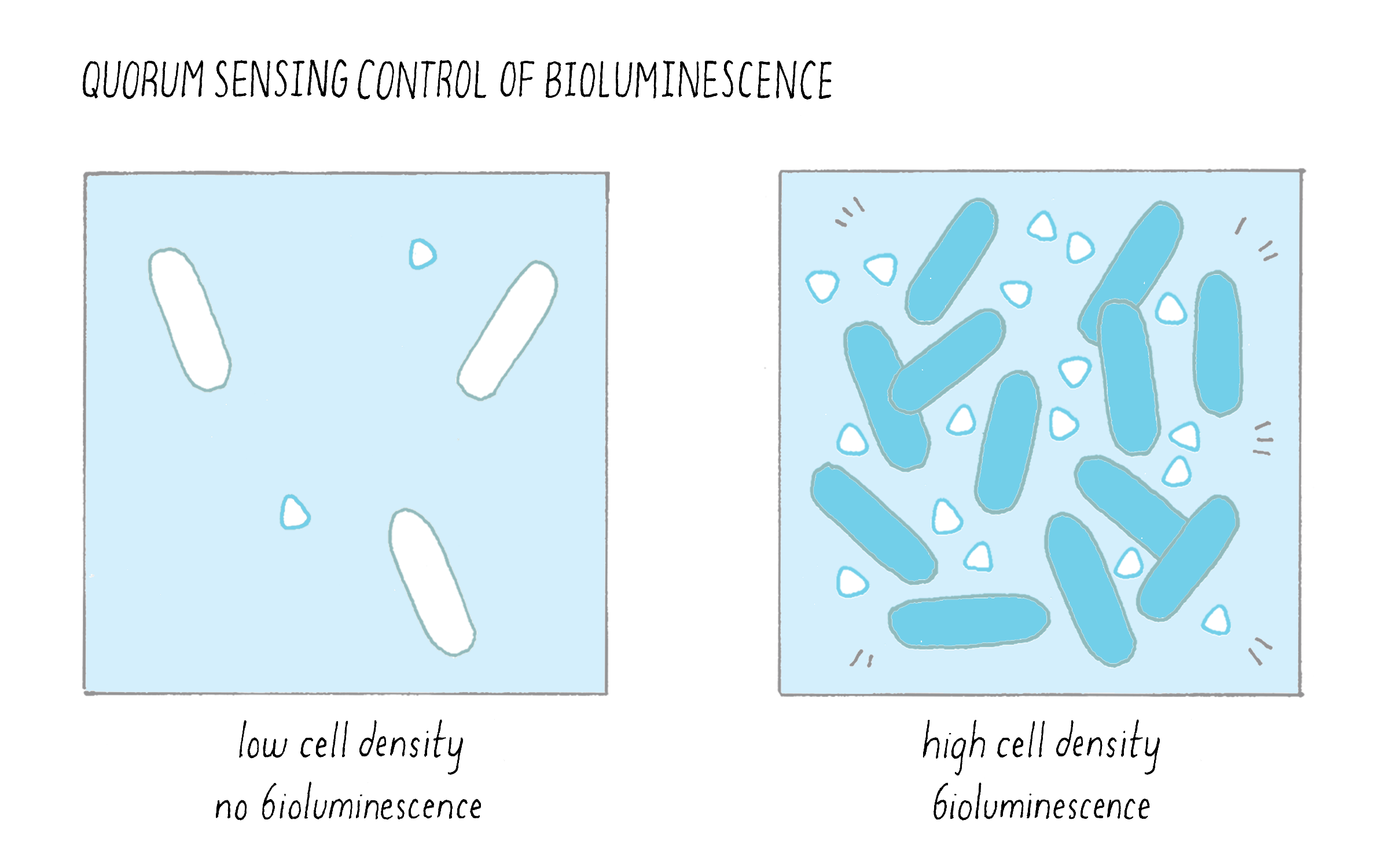
1) Bacteria produce and release small molecules, called autoinducers, into the extracellular environment: At low cell density, when only a few bacterial cells are present, only a small amount of autoinducer can be made and it diffuses away. Because the autoinducer concentration is low, the bacteria cannot detect it. Under this condition, the bacteria act as individuals. As the cells divide and increase in number, they produce and release more autoinducer molecules into the environment; the extracellular concentration of autoinducer increases in step with increasing cell-population-density. This principle is illustrated in Figure 11 in the context of bioluminescence.
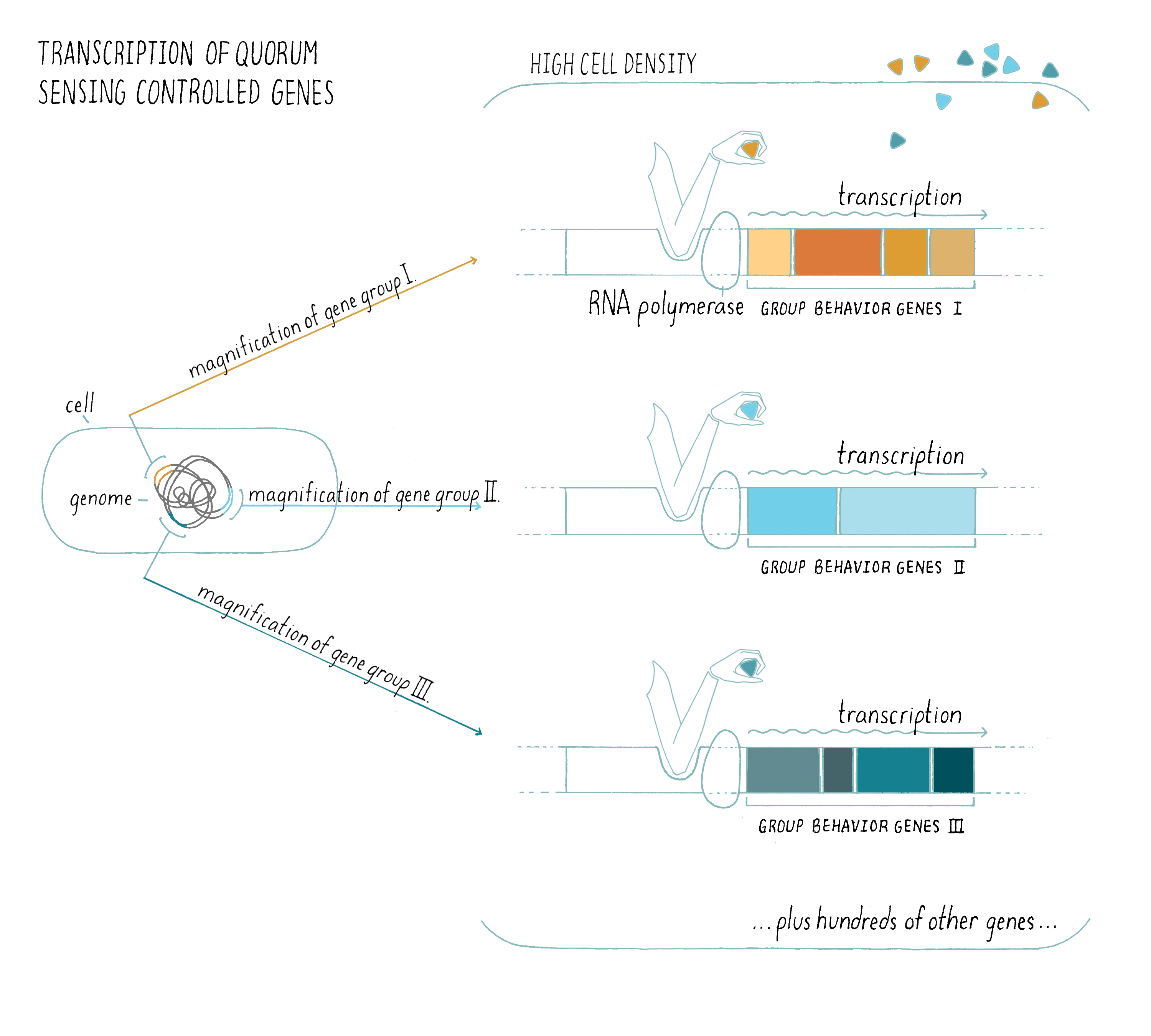
2) Typically, autoinducers are freely diffusible across the cell membrane, so the extracellular concentration is equal to the intracellular concentration. When the extracellular autoinducer reaches a critical threshold concentration, which is indicative of a particular cell population density, the bacteria detect the accumulated autoinducer using receptors (either in the cytoplasm or in the plasma membrane). In response, autoinducer-bound receptors regulate genes that underlie collective behaviors. Regulation involves the precise control of transcription factors that interact with RNA polymerase and this determines whether a gene will be transcribed or not. The types of genes being turned “on” or “off” by quorum sensing give the bacteria distinct behaviors or phenotypes in the low cell density and high cell density situations (Figure 12). For example, quorum-sensing systems can control genes enabling biofilm formation, virulence factor production, antibiotic production, symbiosis, DNA uptake, etc. Typically, using quorum sensing, a given bacterial species will control between 200 and 600 genes (out of ~2,000–4,000 total genes) which are encoded at many locations in the bacterial genome, so a large proportion of a bacterium’s genome can be devoted to group behaviors (Figure 12).
Quorum Sensing Involves Signaling Pathways
Accumulated autoinducers are detected in one of two ways:
1) by cytoplasmic receptors or
2) by plasma-membrane-bound receptors
It is noteworthy to mention here that many eukaryotic signal transduction systems (such as steroid hormones [cytoplasmic receptor] and insulin [membrane receptor]) work analogously to these two types of bacterial quorum-sensing signal transduction systems.
Cytoplasmic Receptors as Transcription Factors
The simplest quorum-sensing systems to understand are the ones like those discovered by Engebrecht and Silverman in V. fischeri. Accumulated, extracellular autoinducer diffuses inside of the cell and binds to the cytoplasmic receptor (called LuxR). The cytoplasmic receptor is a transcription factor, which means that it binds to a specific promoter DNA sequence. This system acts as a switch: the cytoplasmic receptor will only bind to the DNA when it is bound to its autoinducer (Figure 13). Once the autoinducer-bound cytoplasmic receptor binds the DNA, it then recruits and activates RNA polymerase to also bind the DNA. RNA polymerase then starts transcribing messenger RNA (mRNA) encoding the nearby genes. The mRNA, in turn, instructs ribosomes to synthesize the proteins encoded by the genes (see Central Dogma). In our example of bioluminescence, the expressed genes encode the light-producing luciferase enzyme and the LuxI enzyme that makes the autoinducer.
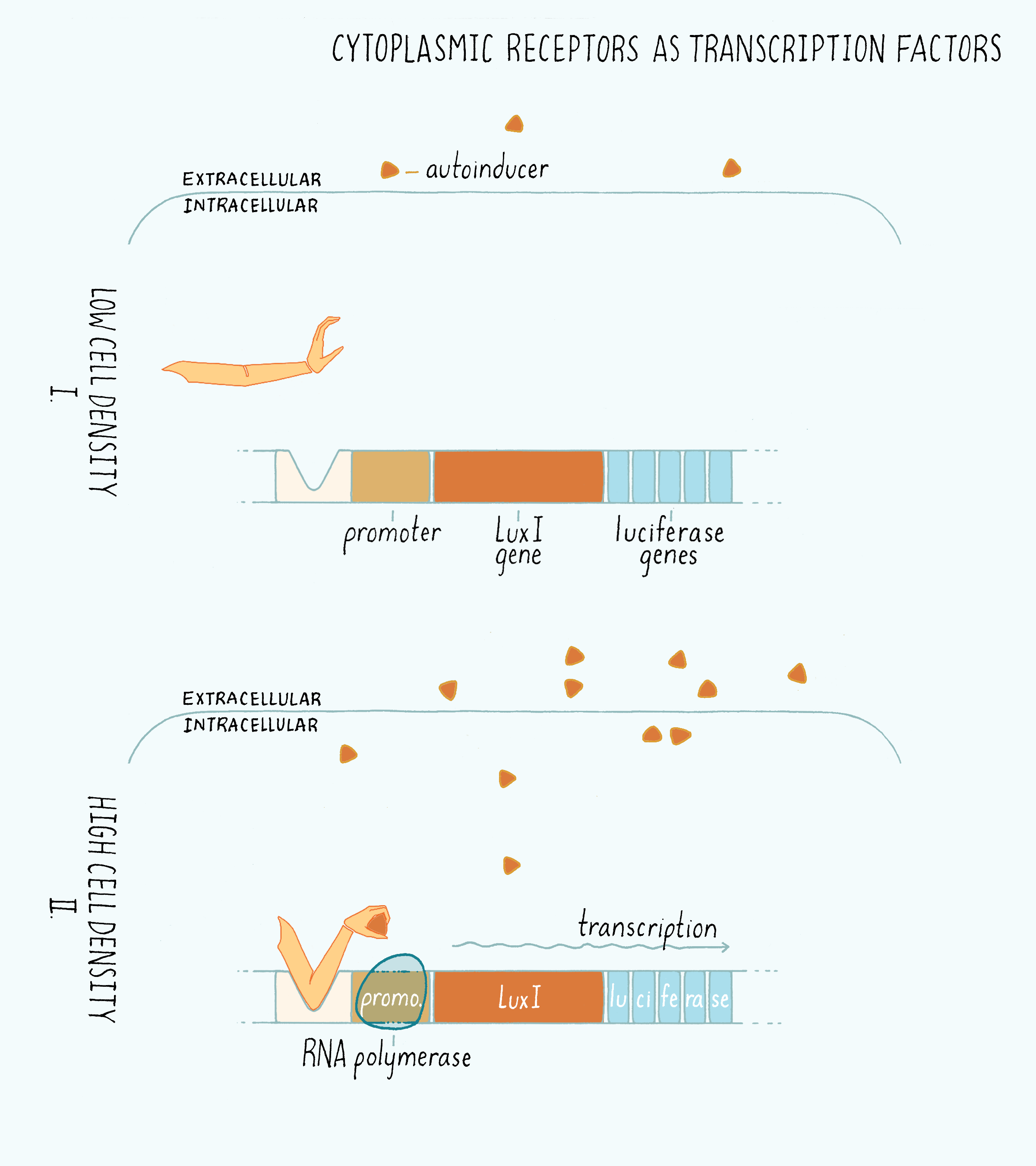
Membrane-Spanning Receptors and Phosphorylation Cascades
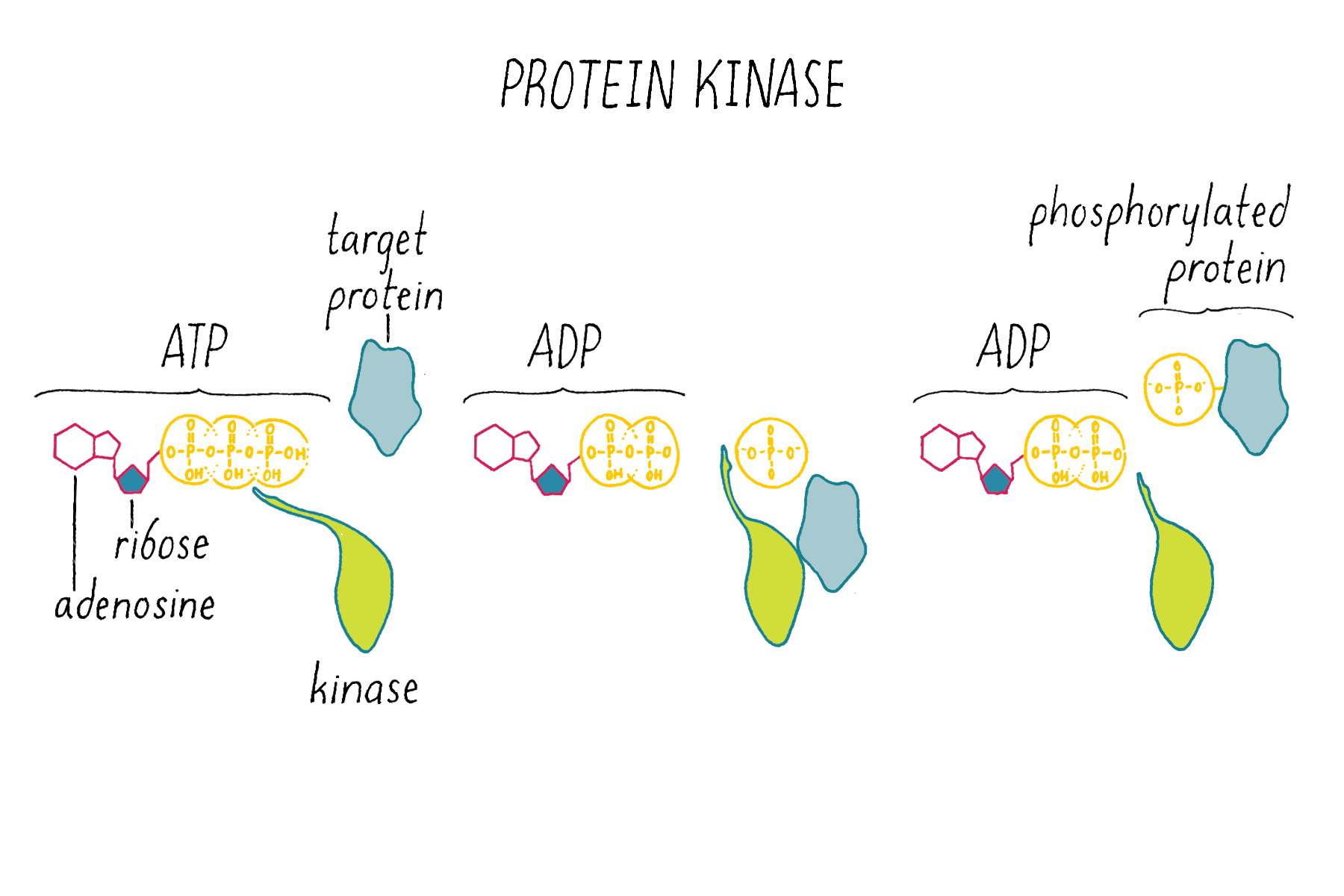
Many quorum-sensing systems involve plasma-membrane-bound receptor proteins. The domain of the receptor that is located on the outside of the cell possesses a pocket that binds a specific autoinducer. The domain of the receptor protein that resides on the inside of the cell is an enzyme with two activities called kinase and phosphatase.
Kinases (Figure 14) are enzymes that, using ATP as a chemical substrate, covalently link a phosphate to a particular amino acid on a protein. The addition of the phosphate to the protein serves a regulatory function; it changes the activity of the protein.
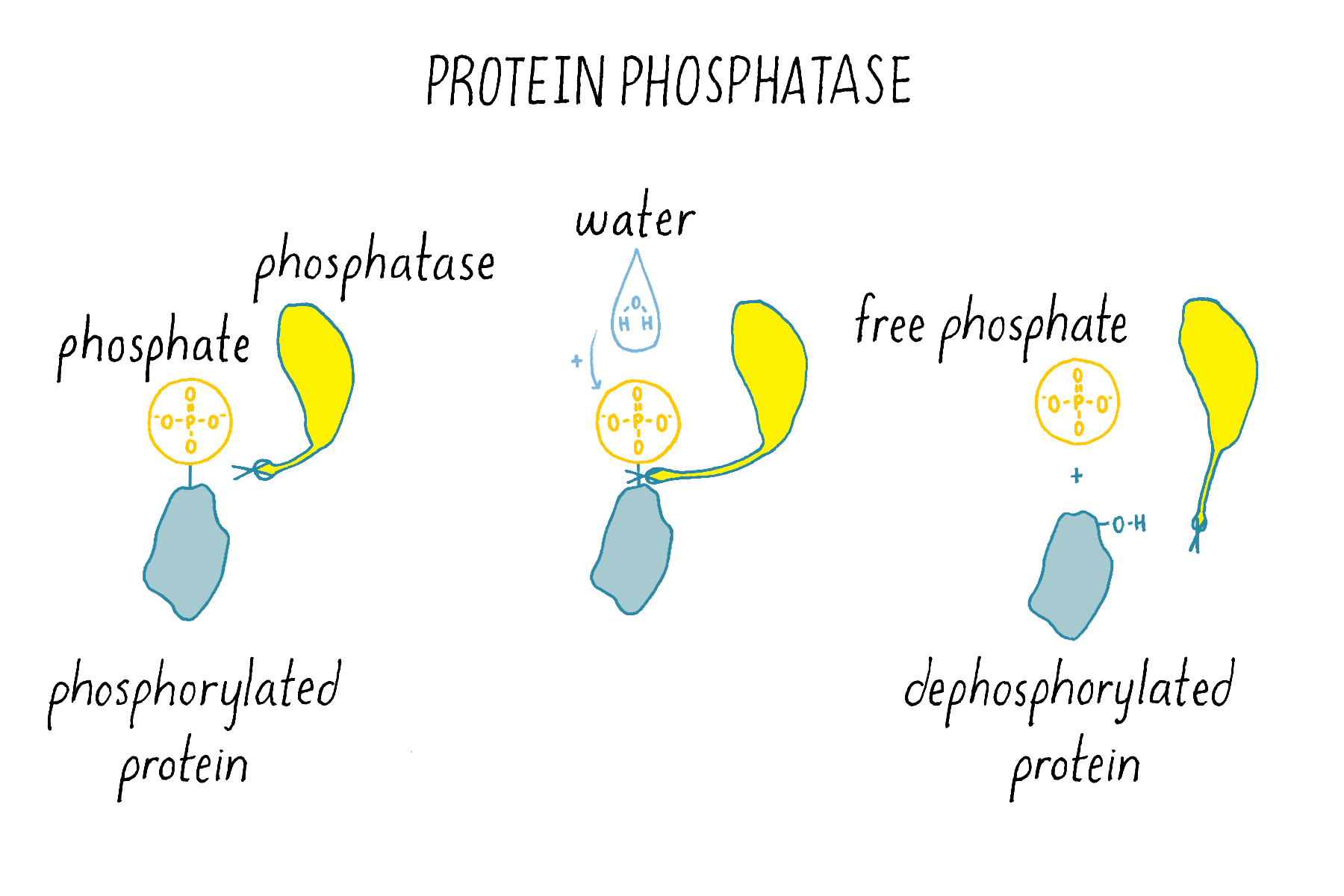
Phosphatases (Figure 15) are enzymes that remove phosphate from a protein. Thus, kinases and phosphatases have opposing enzymatic activities.
In the case of quorum sensing, at low cell density, when the autoinducer has not accumulated, its partner plasma-membrane-bound receptor acts as a kinase and phosphorylates itself (Figure 16). That phosphate group is shuttled through a series of other intermediary proteins, ending with the transfer of the phosphate to a transcription factor. That phosphorylated transcription factor, together with RNA polymerase, causes genes that are needed for the bacteria to act as individuals to be transcribed while genes needed for group behaviors are not transcribed (Figure 16).
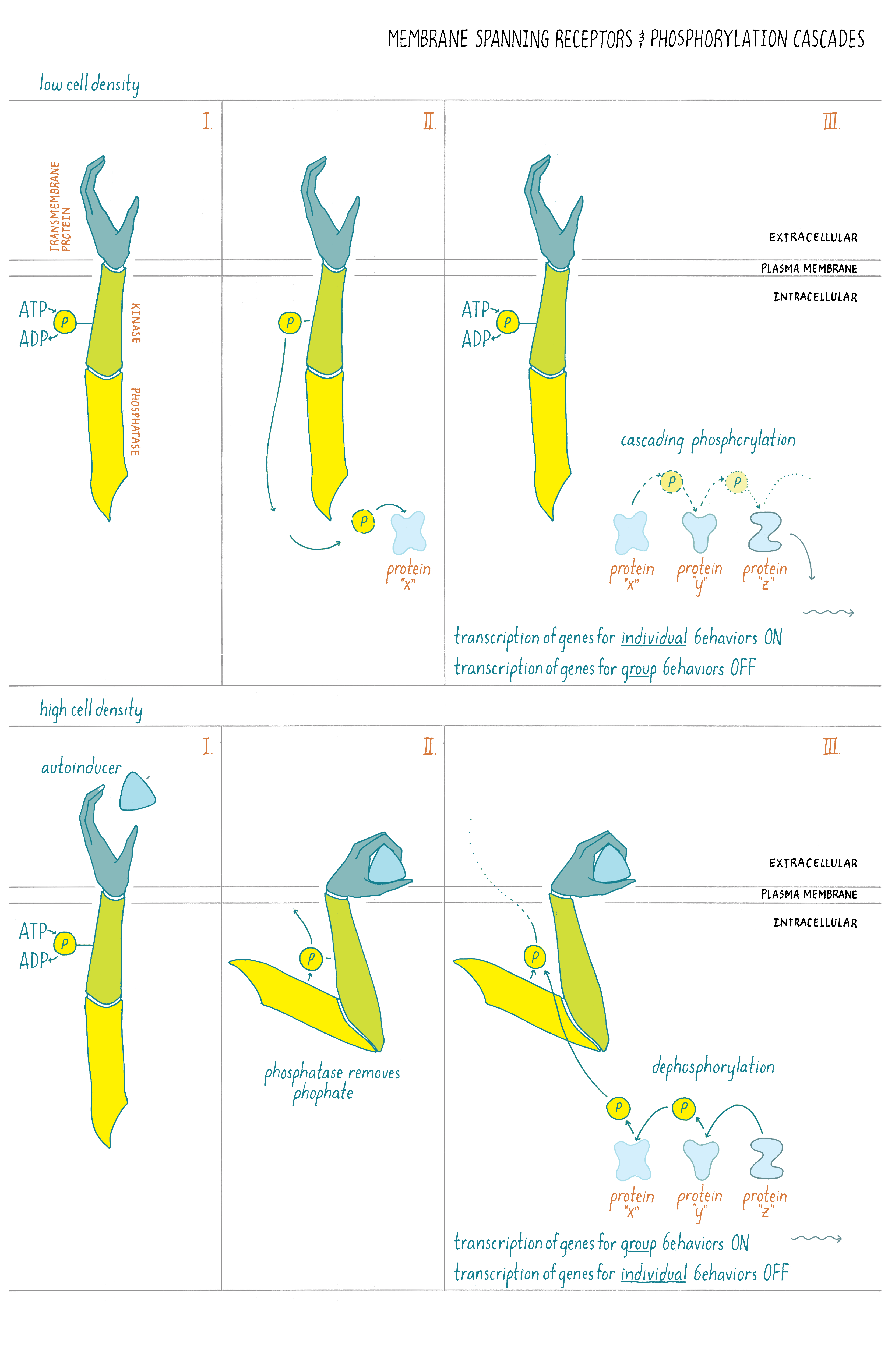
What happens at high cell density? The autoinducer levels increase, enabling autoinducer molecules to bind to the plasma-membrane-bound receptor. Autoinducer binding changes the receptor from being a kinase to being a phosphatase. The receptor removes the phosphate group from itself. This reverses the flow of phosphate from the transcription factor, through the intermediate proteins, to the receptor and ultimately these backward-flowing phosphates are removed from the receptor. Under this condition, genes required for individual behaviors are not transcribed while genes required for group behaviors are transcribed into mRNA and then made into proteins. (Figure 16).
The role of the positive-feedback loop
Quorum-sensing systems also illustrate a prevalent control system in biology called “feedback loops.” If you study how cells make decisions, you will encounter feedback loops over and over again. Feedback loops are not unique to biology. Electrical engineers use feedback frequently in circuit design. A concise definition of positive feedback can be found in Wikipedia: “Positive feedback is a process that occurs in a feedback loop in which the effects of a small disturbance on a system include an increase in the magnitude of the perturbation. That is, A produces more of B which in turn produces more of A.”
Let us see how this principle plays out in quorum sensing. In Figure 17, the input is the autoinducer, which triggers “activity A,” transcription of genes. Gene transcription generates a desired output, such as bioluminescence. However, it also generates more of the autoinducer production enzyme, which acts as “activity “B.” The enzyme activity then results in more input (the autoinducer), which results in more transcription (“activity A”).
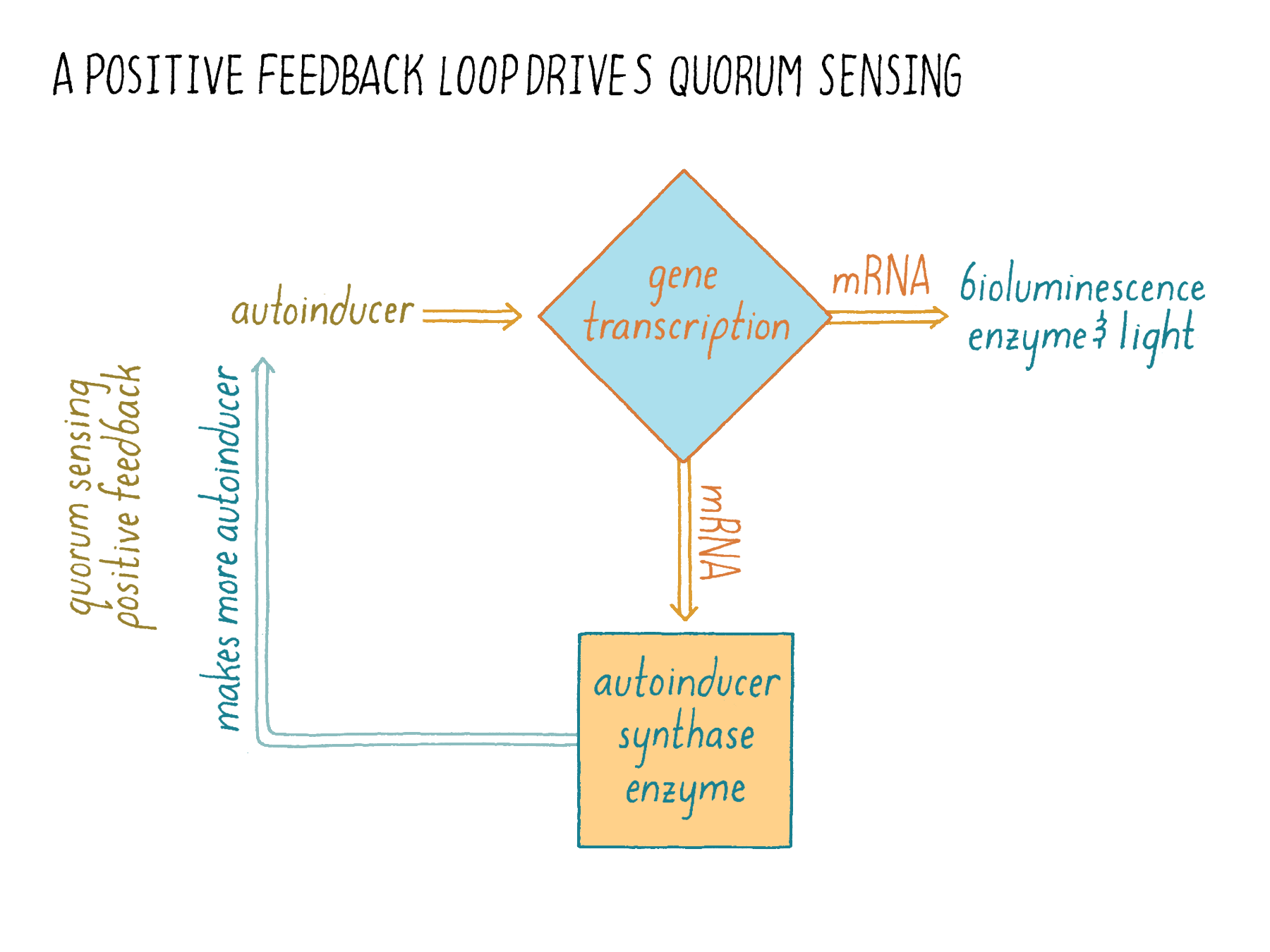
What does positive feedback do? In this case, it causes a switch-like change in the behavior of the bacterial population. Once a certain level of autoinducer is reached (due to bacterial cell growth), the production of autoinducer suddenly increases rapidly because more of the autoinducer production enzyme is made. This “bolus-dose” of autoinducer encourages all of the nearby cells to switch into quorum-sensing mode presumably promoting commitment to and synchrony in the execution of group behaviors. Without positive feedback on autoinducer production, you can imagine that, overall, cells would experience only slowly increasing autoinducer concentrations making the transition sluggish. Such a communication system would be noisy and inefficient, with some cells detecting enough autoinducer to engage in quorum sensing while others that encounter less autoinducer, lagging behind in the quorum-sensing transition. It is this positive feedback loop in which the production of the communication molecule is activated by the quorum-sensing relay that led to the name “autoinducer.”
Intra-species, Intra-genera, and Inter-species Communication
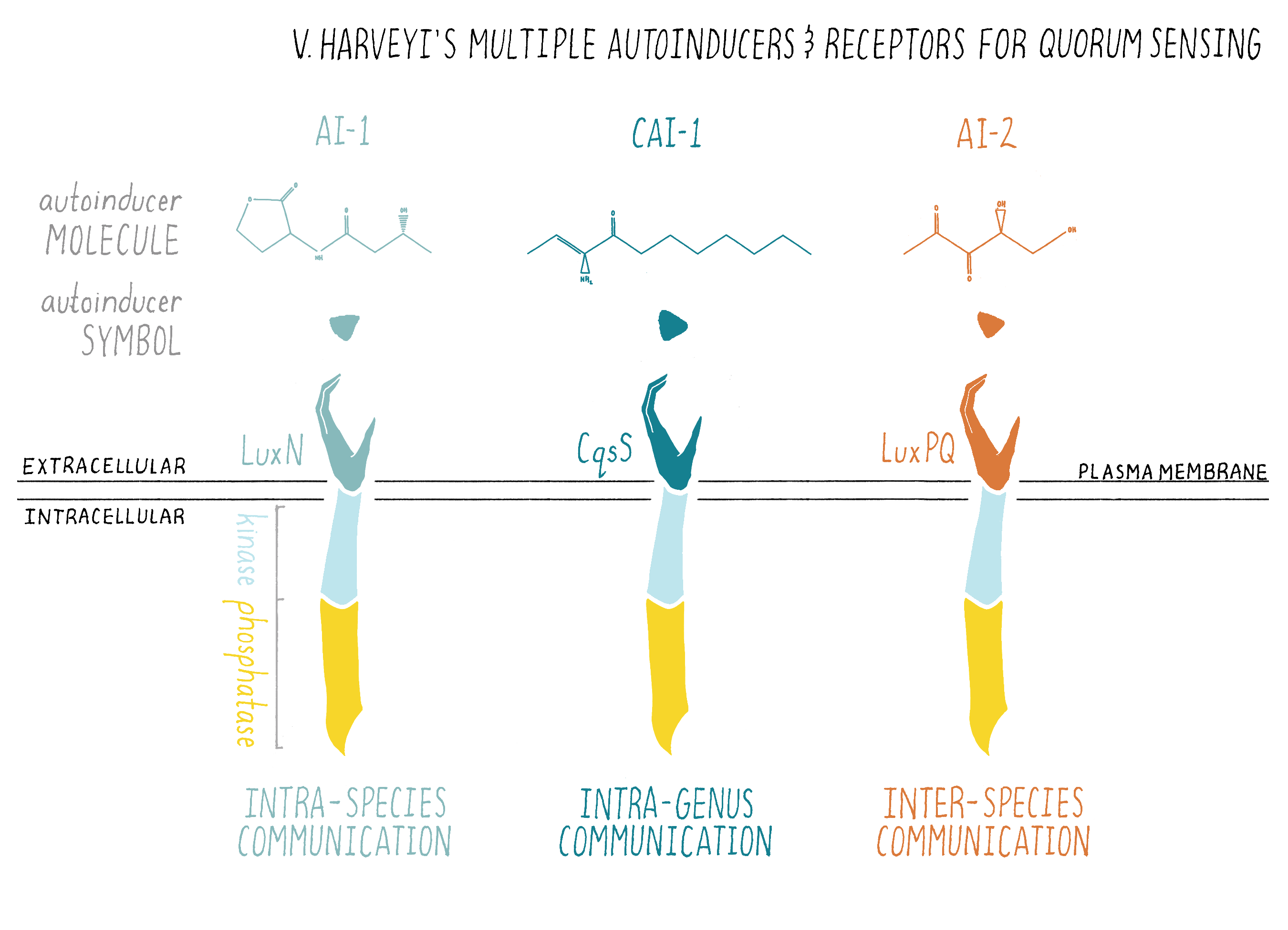
Bacteria often use multiple autoinducers for quorum sensing and the chemical nature of each autoinducer encodes different information about “who” is in the vicinity. Take V. harveyi: it uses three different autoinducer chemicals for intra-species, intra-genera, and inter-species communication, respectively (Figures 10 and 18). Only V. harveyi and its closest relative produce the chemical AI-1, suggesting that the AI-1 autoinducer is used for intraspecies communication. AI-1 is a homoserine lactone, similar to the homoserine lactone identified originally as the V. fischeri autoinducer. This is a common theme in quorum sensing: distinct homoserine lactone compounds, i.e., molecules that are similar but not identical, are used by different bacteria for intraspecies communication. Vibrios (all bacteria of the genus Vibrio) make another chemical called CAI-1, suggesting that CAI-1 is used for intra-genus communication. By contrast, the autoinducer chemical called AI-2 is broadly made among bacteria, so AI-2 is proposed to function in interspecies communication. V. harveyi has the ability to determine the ratio of AI-1:CAI-1:AI-2.
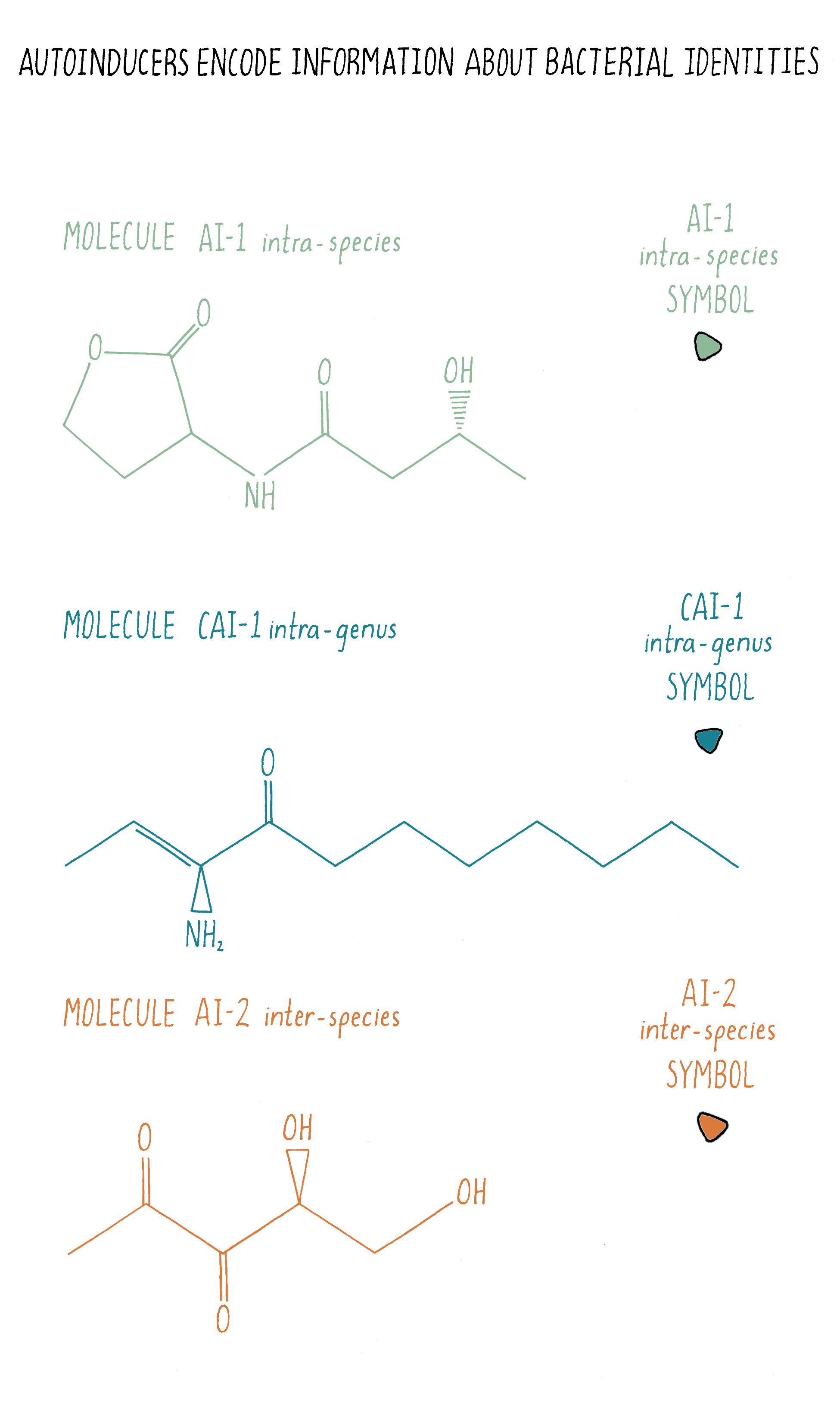
Interestingly, V. harveyi tailors its quorum-sensing-controlled behaviors based on which autoinducer signal dominates. Presumably, V. harveyi decodes the information in the relative concentrations of the three autoinducers to determine whether it and its relatives or, alternatively, some other bacterial species is in the majority of a mixed-species community. On the basis of that information, V. harveyi carries out quorum-sensing behaviors that are appropriate to its circumstances. This means that, through quorum sensing, bacteria can tell self from non-self!
Who is making all these different autoinducers? In the above explanations concerning discoveries of components involved in quorum sensing, the bacteria under study were grown as single species in flasks in laboratories. This initial strategy was sensible for doing simple experiments to discover the basic principles of quorum sensing. However, in nature, bacteria live in complex and changing environments, in biofilms, and frequently, they live in communities with other bacterial species present.
Take for example the human microbiome, which I mentioned earlier in this Narrative. Your mouth, for example, is a thriving community of many bacteria that live in biofilms on your teeth and gums. There are also very complex bacterial communities in the human gut, estimated at 1014 bacterial cells (more than the number of human cells in your body!) made up of 600 different species, primarily growing in biofilms. You can imagine that many autoinducers are present, that their levels vary (there is periodic fluid flow in the gut that could concentrate or wash away autoinducers) and changes in nutrients (depending on when and what a person eats) can allow particular bacteria to grow faster or slower at different times. Somehow, the individual bacteria in these diverse consortia have to be able to measure and interpret autoinducer information to discern how many and who their neighbors are so they can behave appropriately. Moreover, they have to be able to update their assessments to deal with changes in community composition.
Scientists do not yet understand much about how quorum sensing works in the human microbiome (or any complicated mixed-species bacterial community) because it is early days in such studies. Here we, nonetheless, try to give insight into some possibilities based on what we do know from the findings discussed above. Figure 19 provides a picture of how we can begin to think about quorum sensing in communities containing multiple species of bacteria. For simplicity, we only consider intraspecies and interspecies autoinducers. The top panel of Figure 19 shows the mixed-species community. Imagine many different species of bacteria residing in close proximity to one another. In the Figure, each color pattern represents one bacterial species.

Explorer’s Question: What information do you think the “yellow bacteria” in our cartoon can glean about their environment?
Answer: The lower left panel highlights the “yellow” species. In our scheme, there are many “yellow” bacteria in the community. They encounter a high level of their own intra-species autoinducer (yellow triangles) as well as a high concentration of the inter-species autoinducer (blue triangles). That blend of autoinducer tells the “yellow” species that they are at high cell density but they have neighbors that are non-kin.
Explorer’s Question: Take the “orange” species in the bottom right panel. What information do you think that lone orange bacterium infers from the autoinducer blend?
Answer: In our cartoon, there are only a few bacteria of the “orange” species present (look at the original, top panel). The “orange” bacterial species detects none or only a very low level of its species-specific autoinducer (orange triangles) but it encounters a high level of the interspecies autoinducer (blue triangles). That combination of autoinducers tells the “orange” species that it and its kin are far outnumbered by non-kin.
Going back to the top panel, in our scenario, the “yellow” species would likely turn on many quorum-sensing-controlled genes that help it prosper as a group. However, the “yellow” species might not turn on its entire repertoire of quorum-sensing-controlled genes because it would have detected that there also are many non-kin neighbors around that could compete for and take advantage of released molecules. The “yellow” species knows there are non-kin present due to the detection of the inter-species autoinducer. It might activate defensive behaviors to ensure it maintains the majority in the community.
The “orange” species would not launch its quorum-sensing program because there are not enough members of its kin to productively act as a group. The “orange” species would likely wait and keep monitoring the situation until such time that it detects a higher level of its species-specific autoinducer (orange triangles). Such a change in the ratio of the different autoinducer molecules could alert the “orange” species that it was gaining in cell numbers in the mixed-species community.
Part III: Frontiers —
Quorum Sensing and Disease
In this section, I would like to share what I believe are two particularly exciting frontiers for quorum sensing.
First, scientists are developing strategies to manipulate quorum sensing, which holds promise as new means for treating bacterial infections or for promoting beneficial bacteria.
A second frontier is understanding how quorum sensing operates in the “wild”, rather than in the laboratory. In the Journey to Discovery, scientists studied quorum sensing of a single bacterial species growing in a nutrient-rich medium in a flask. In nature, however, bacteria grow in complex environments that include many bacterial species, viruses, and often eukaryote hosts (such as the inside of our gut) and the bacteria live in changing nutrient conditions. Understanding quorum sensing in such complex settings may enable us to better understand and treat disease (Video 3). The cholera disease is discussed as such an example.
Quorum-Sensing Interference Strategies
Often, bacteria use quorum sensing to cause disease. If we understand the language, can we interfere with quorum sensing to halt virulence (a kind of cyber warfare against the bugs)? Scientists are now using what they know about autoinducer structures and how autoinducers interact with their partner receptors to make synthetic molecules that act as antagonists: molecules that bind to autoinducer producing enzymes or to the autoinducer receptors and inhibit their functions. In so doing, such molecules suppress quorum-sensing behaviors, including virulence.
In addition to thinking up strategies from scratch, scientists often look to nature itself for inspiration. As it turns out, some bacteria have evolved strategies for interfering with each other’s quorum-sensing communication circuits. Here are some examples:
- Certain bacteria secrete enzymes that can clip a competitor’s autoinducer in half, making it inactive.
- Other bacteria ravenously consume their competitor’s autoinducers, using the chemical building blocks in the autoinducer (carbon, nitrogen) as nutrients to help the consumer to grow.
In both of the above cases, because the autoinducer disappears, the autoinducer-producing bacteria miscount and do not properly execute their quorum-sensing-controlled behaviors. Scientists are seeking to understand these naturally occurring instances of quorum-sensing interference and then re-deploy them to manipulate quorum sensing on-demand as highlighted in the example below.
Certain species of Bacillus secrete an enzyme (called a lactonase) that cleaves the carbon rings of homoserine lactone autoinducers produced by other bacteria. This action renders the autoinducer-producing bacterium “speechless,” that is, incapable of quorum-sensing-mediated communication and, in turn, incapable of quorum-sensing-driven group behaviors. Bacillus bacteria do not rely on homoserine lactones as autoinducers, so the Bacillus quorum-sensing system is impervious to the cleaving enzyme. Thus, Bacillus, while shutting down its neighbor’s quorum-sensing traits, remains capable of properly enacting its own quorum-sensing behaviors.
The originally discovered Bacillus lactonase enzyme (called AiiA) and many homologs have been intensively studied and exploited by scientists to quench quorum sensing in a variety of scenarios. For example, Dong et al. introduced the gene that encodes the AiiA enzyme into plants to make the plants resistant to bacterial pathogens that require quorum sensing to cause disease (see the Narrative on Plant Genetics by Ronald to learn more about how to make transgenic plants). The left side of Figure 20 shows slices from potatoes that were infected with a bacterial pathogen, and the plant exhibits evidence of massive infection. In contrast, slices from a plant that is producing the AiiA enzyme from the introduced gene remain largely free of bacterial infection (right side of Figure 20).
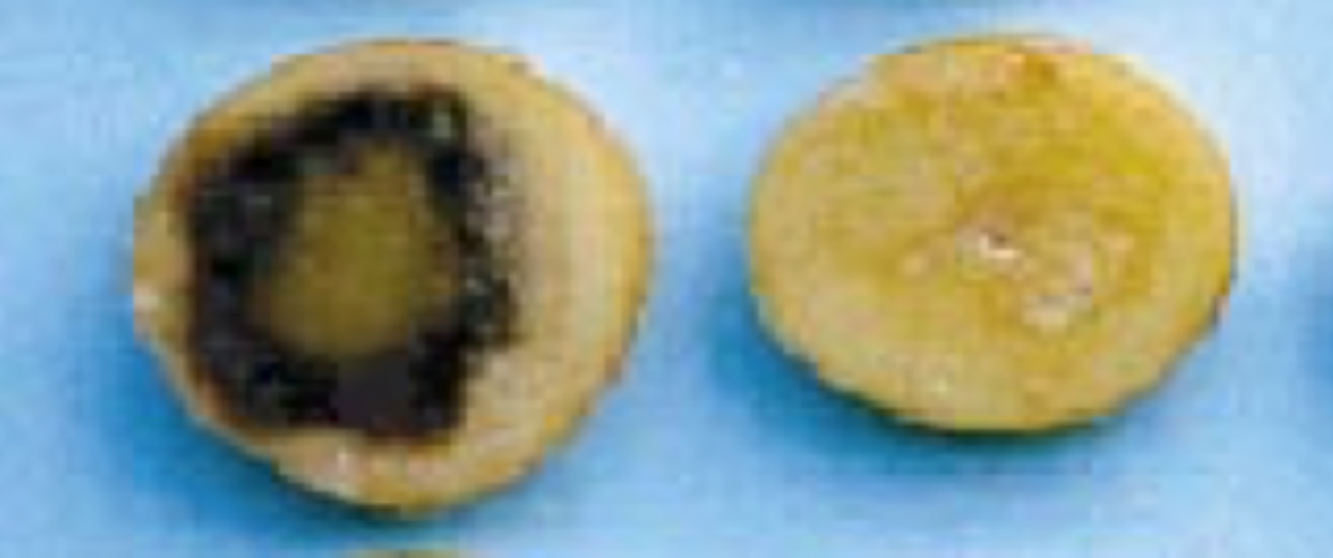
Another way scientists are trying to exploit AiiA to reduce infections in plants is to supply a beneficial bacterium that naturally makes an AiiA-type cleaving enzyme to the soil. Bacillus thuringiensis is such a bacterium and, by degrading autoinducers, it keeps other, harmful bacteria from infecting plant roots. Scientists found that significantly less disease occurred in plants if AiiA-producing B. thuringiensis was included in incubations of potato tubers with pathogenic bacteria. Importantly, they also showed that rotting could be stopped when an AiiA-producing bacterial strain was added to the soil even 2 days after infection of the potato plant with the pathogen. This last result suggests that strategies with AiiA, beyond being preventative, could be curative. Many other quorum-quenching applications for AiiA have been developed since this original work.
Explorer’s Question: Can you think of other strategies to interfere with quorum sensing?
Answer: The following strategies are currently being pursued: (1) Make autoinducer antagonists -compounds that are similar in structure to the real autoinducers so they are capable of binding the partner receptors, but, unlike the autoinducers, antagonists prevent the receptors from activating quorum-sensing-controlled gene expression. (2) Make compounds that inhibit the autoinducer production enzymes, so that no autoinducers are produced. (3) Make antibodies that bind and sequester autoinducers so the autoinducers are not available to interact with the partner receptors. In all three cases, the idea is that quorum sensing will not commence, virulence genes will not turn on, and the pathogen will be rendered non-virulent and susceptible to clearing by the immune system. (4) Make potent pro-quorum-sensing compounds. Such molecules, called agonists, are basically super-autoinducers. Agonists would cause quorum sensing to be activated prematurely so the pathogenic bacteria would turn on virulence genes before sufficient numbers of bacterial cells are present to mount a productive infection. Premature quorum sensing should also alert the host immune system to come wipe out the pathogens.
The above example illustrates how disrupting quorum sensing could be useful for agriculture. However, the AiiA strategy, as well as the strategies you and I came up with, have uses far beyond plants. Notably, many quorum-sensing interference strategies are being harnessed to fight infectious diseases in humans. Some of these quorum-sensing interference strategies are wholly “synthetic”, that is, scientists use what they know about quorum sensing to develop interference methods from scratch (antibodies, autoinducer antagonists, autoinducer production enzyme inhibitors, etc.). In other cases, similar to how AiiA was discovered and used, scientists are taking hints from nature to come up with quorum-sensing disruption methods.
Quorum-Sensing in Natural Contexts
Beyond studying bioluminescent V. harveyi, my group also studies its pathogenic relative, Vibrio cholerae. V. cholerae causes the diarrheal disease cholera (Video 4). People get cholera from eating contaminated foods or from drinking contaminated water. Cholera is endemic in different regions of the world where it causes ~4 million cases and ~140,000 deaths per year. V. cholerae requires quorum sensing to be a pathogen. We are using our V. cholerae studies to learn how quorum-sensing operates in the human host—that is, in the gut, which has topography (many curves, winding paths, grooves, and edges). The gut also has episodic flow with gradients, pulses, eddies, and currents, and a changing nutrient supply. In addition to its human cells, the gut is home to a microbiome containing 1014 microbial cells comprised of 600 different species, each with their own unique biology, and, in many cases, making their own unique quorum-sensing autoinducers.
We wondered how V. cholerae uses quorum sensing for launching and terminating its virulence program in the complex native environment of the gut? And, might it be possible to manipulate quorum sensing to treat or prevent this infectious disease (Video 4)?
Here is what we have learned so far:
Unlike most quorum-sensing bacteria studied to date, high autoinducer concentrations repress (rather than activate) virulence genes and biofilm formation in V. cholerae. Furthermore, autoinducers activate genes required for V. cholerae cells to disperse from the host. The way we interpret these findings is as follows:
- V. cholerae causes an acute disease. Its strategy is to get into the host, multiply greatly, and get out of the host in huge numbers to go on to infect the next person.
- A person ingests V. cholerae from contaminated water. It enters the human host at low cell density with its repertoire of biofilm and virulence genes highly expressed; this enables the pathogen to adhere to the host intestinal cells and cause infection.
- As the V. cholerae cells grow and divide in the human intestine, their autoinducers accumulate. When the bacteria detect the buildup of autoinducers, quorum-sensing signal transduction causes repression of V. cholerae virulence and biofilm genes and activation of dispersal genes. What ensues is the terrible diarrhea characteristic of cholera disease.
- V. cholerae uses the watery diarrhea as a means to escape from the host, to disseminate into the environment at high numbers, presumably to infect the next human.
Explorer’s Question: In V. cholerae, autoinducers repress virulence and biofilm formation and they promote dispersal. How might you use autoinducer interference strategies to combat cholera disease at different times?
Answer: One can imagine that people who are prone to getting cholera disease could be prospectively treated with an autoinducer agonist as a preventative measure. In this scenario, if the person got infected with V. cholerae, the presence of the autoinducer agonist would drive V. cholerae into the non-pathogenic, dispersal mode. The bacteria would leave the host without multiplying and without causing disease. Another possibility is to treat people who have cholera disease with an autoinducer agonist, in this case, prematurely forcing V. cholerea to disperse and thereby reducing disease severity (such a strategy would need to be developed hand-in-hand with public health officials because it could have the consequence of increasing the spread of the disease through the dissemination of the bacteria out into the environment).
The V. cholerae quorum-sensing signal transduction pathway is similar to that of V. harveyi except that it only has two autoinducers (CAI-1 and AI-2) and their partner receptors (see Figures 10, 18, and 19). In our effort to understand the role of V. cholerae quorum sensing in natural contexts, we recently discovered a new autoinducer called DPO. Our discovery occurred when two members of the lab (Kai Papenfort and Justin Silpe) found a V. cholerae gene (vqmR) that was not expressed when V. cholerae cells were at low cell density but was expressed when the cells were at high cell density—the hallmark quorum-sensing pattern.
To test if autoinducers controlled vqmR expression, Kai and Justin took cell-free conditioned medium from high-cell-density V. cholerae cells and added it to a culture of low cell density V. cholerae cells and, after making the addition, they measured vqmR levels in the culture.
Explorer’s Question: What would you expect would happen to the expression of vqmR in the experiment above? Would it go up, down, or stay the same?
Answer: It would go up because the low-density cells were “tricked” into thinking that there were in a high-density environment. This is the same approach that Ken Nealson took in the original discovery of the V. fischeri quorum-sensing autoinducer (Figure 6).
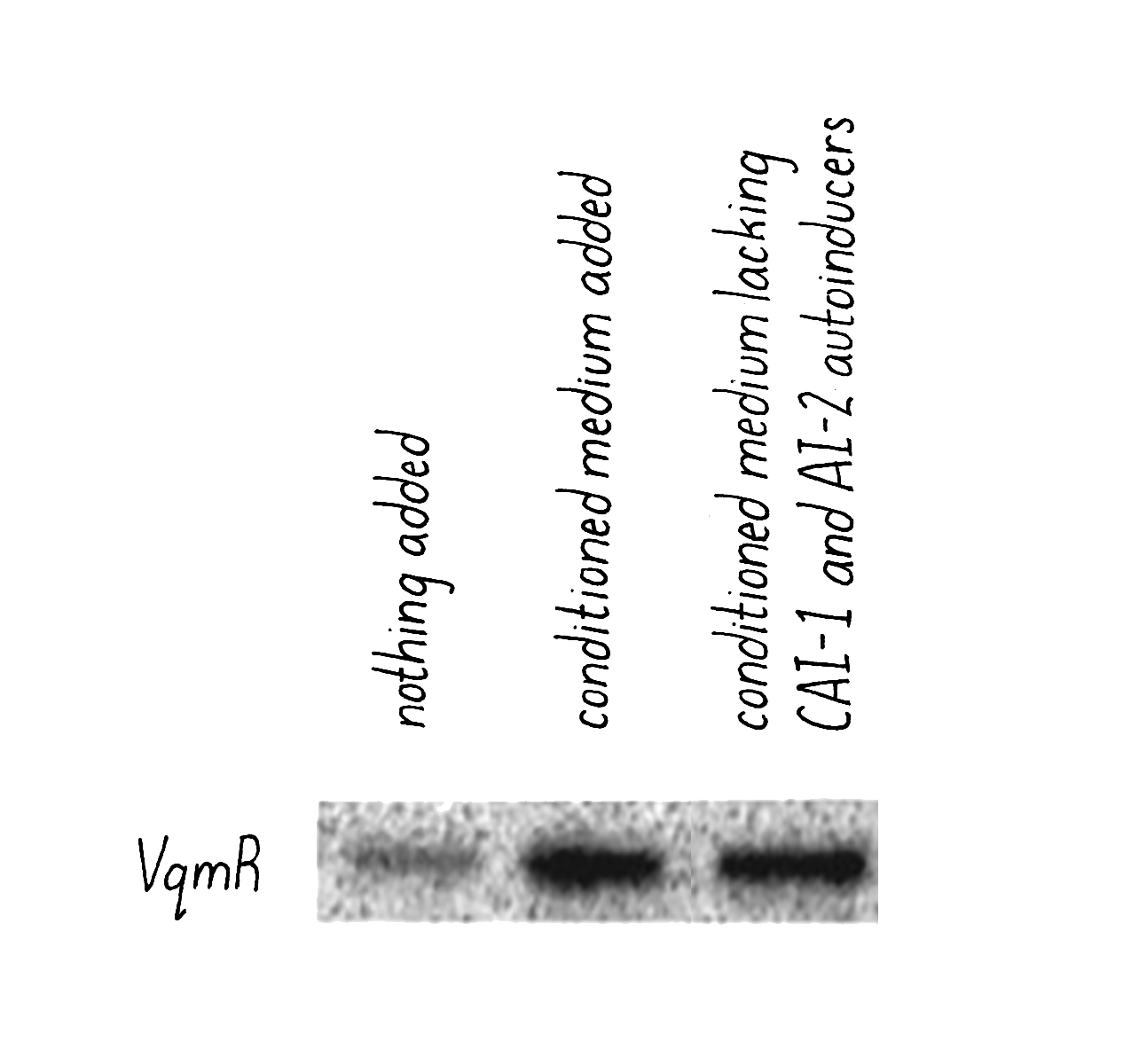
Justin and Kai also added conditioned medium prepared from a V. cholerae mutant strain that was incapable of producing the known autoinducers CAI-1 and AI-2. They did this addition as a control experiment to show that autoinducers were involved in the response. They expected that nothing would happen- vqmR mRNA levels would remain unchanged, as shown through a method called a Northern Blot (Dig Deeper 4). But science can be full of surprises. What they found is that the conditioned medium lacking the known autoinducers still induced increased expression of vqmR mRNA (Figure 21). That surprising result meant that there must be some additional activating substance other than the known autoinducers CAI-1 and AI-2.
Kai and Justin then went on a hunt to find this new mystery autoinducer (Video 5). Using traditional fractionation techniques, together with chemical identification technologies, they identified the new autoinducer and named it DPO (shorthand for a longer chemical name). In follow-up biological studies, Justin and Kai found that DPO is produced by V. cholerae as well as by some members of the normal human gut microbiome. Bacteria synthesize DPO by linking together parts of two amino acids, threonine and alanine. The human protein mucin, which coats our intestinal cells, is rich in threonine. Thus, we think that gut microbiome bacteria are like farmers; they harvest human-made mucin to obtain one of the building blocks needed to make DPO.
We think that V. cholerae assesses DPO levels to determine when to complete its pathogenic cycle: DPO suppresses the V. cholerae virulence program, terminates biofilm formation, and activates dispersal genes. We think that DPO made by human microbiome bacteria limits V. cholerae infection by causing V. cholerae to disperse prematurely. Indeed, another group of scientists showed that in a mouse model of V. cholerae infection, the particular community of gut microbiome bacteria present in the gut dictates how much DPO is made (since some microbiome bacteria can make DPO and some cannot). The idea is, if more DPO is made by the normal gut microbiome, this would limit the severity of the cholera disease.
Thus, interactions across the eukaryotic and bacterial kingdoms rely on at least one shared quorum-sensing autoinducer, DPO. Presumably, each entity (the human host, the beneficial microbiome bacteria, and the pathogen V. cholerae) is reacting to information encoded in the DPO molecule to optimize its survival.
Closing Thoughts
The history of quorum sensing, from my point of view, provides insights into the process of science, which I share with you here.
• Lessons about the need for curiosity-driven research: Many people think that research should always be pragmatic, for example, relevant to medicine. Often, relevance can only be appreciated long after initial discoveries are made, indeed, only when scientists have had time to delve deeply into a subject and they have learned enough to put disparate findings together into a cohesive picture. No one imagined that studying bioluminescence would lead to biotech using those discoveries to develop new antimicrobial medicines, notably in a time when the global problem of antibiotic resistance has reached an epic scale.
• Exciting unanswered questions: In every case to date, the molecules scientists discovered as quorum-sensing autoinducers are brand new molecules to mankind, suggesting that bacteria house a treasure-trove of compounds that await discovery. If so, can the scientists learn enough about the chemical communication code to harness those molecules, or to make their own “designer molecules” to compel bacteria to perform useful tasks or to stop them from undertaking harmful ones on demand?
• The big picture of applications. Recent research strongly hints that quorum sensing can be manipulated, for example, by supplying quorum-sensing inhibitory molecules to prevent bacteria from infecting animals and plants in model studies of disease. Tethering anti-quorum-sensing molecules to surfaces is now being explored to stop bacteria from adhering to implanted medical devices such as stents and catheters and to prevent the buildup of bacteria into highly resilient and antibiotic-resistant biofilm communities in medical and industrial settings. Pro-quorum-sensing molecules that promote beneficial collective bacterial behaviors are also now being investigated for uses in the bioremediation industry, agriculture, and as probiotics. One grand goal on our horizon is to understand enough about bacterial conversations to be able to come up with strategies to “speak” to bacteria in ways that would be useful to humans, their larger hosts.
• What have we missed? I often wonder how I happened upon quorum sensing in V. harveyi and why I made the particular findings that I described in this Narrative. Since I started my independent career, many students and postdocs have come to my lab and they too have made their own particular findings that moved quorum-sensing research forward. When we work together in lab teams, we encourage one another to pursue particular journeys of discovery, often promoting paths that dovetail with our newest findings, rather than inspiring one another to take some other, seemingly, outlandish path.
I try my hardest to encourage the trainees in my lab to be maximally creative and to do the “crazy” experiment. But I do often wonder and I worry whether I am as adventurous as I used to be and whether, by accident, I hold the field back by biasing mentees in my lab with my view. Perhaps some process even more magical than quorum sensing exists in bacteria. I mean, as I explained, bacteria have been studied for over 400 years, how did quorum sensing get missed for so long? Am I likewise missing some other totally amazing feature in bacteria? Probably! I wonder what other, currently unimaginable, traits lurk in bacteria that can turn science or the prospects for medicine and humanity on their heads. I do hope another generation of explorers will go on journeys of discovery in bacteria and I hope they do it soon.
Dig Deeper
Dig Deeper 1: Symbiosis and bioluminescence
One remarkable feature of this partnership is that, when the sun comes up and E. scolopes (squid) prepares to spend the day buried in the sediment, it pumps out the majority of the V. fischeri bacteria from the light organ, leaving only a small number of cells to regrow during the day and re-populate its light organ. This flushing mechanism accomplishes two essential jobs. First, the squid cannot possibly keep on feeding and growing the V. fischeri bacterial culture in the light organ to an unlimited number of cells over its life-span -- that would take too much of the squid’s resources. Neither the squid nor the bacteria would stay healthy if bacterial growth went unchecked. Flushing out most of the bacteria every morning (~95% of the cells are removed) drastically lowers the number of bacteria needing to be fed by the nutrients in the organ, making this system "manageable" and refreshed daily. Second, as you are learning from the main Narrative, V. fischeri only makes light at high cell density when autoinducers have accumulated and they activate quorum-sensing-controlled light production. The squid only needs the light at night for its "invisibility cloak" effect. Flushing out most of the bacteria in the morning terminates their light production because the flushing mechanism dilutes the bacteria to low cell density and, under this condition, the autoinducer molecules are also flushed out and so they fall to levels below that required for the bacteria to detect them. That is okay, because during the day, the squid is asleep in the sediment and no light is needed. In the light organ, the remaining V. fischeri bacteria replicate as the day goes by, increasing in cell density, they make autoinducer, and it likewise accumulates. And, voila! At night when the squid emerges, conditions in the light organ are such that the cells are at high cell density, high levels of autoinducer are present, the bacteria detect the autoinducer, and they commence light production, exactly when the squid needs the light to protect itself.
Dig Deeper 2 : Bonnie Bassler’s path to becoming a postdoc with Mike Silverman
I "found" Mike Silverman when we both attended the same scientific conference. At that time, I was a Ph.D. student training as a biochemist. For most of the conference, I was bored and did not understand what the speakers were talking about. Then, Mike Silverman gave a talk on his V. fischeri discoveries, which concluded with a figure (similar to the one in Figure 13) summing up his understanding of density-dependent regulation of bioluminescence. I was awestruck. Bacteria could communicate with one another! I remember thinking this is either the most amazing thing I’ve ever heard or that guy is crazy. I decided on the former. Immediately after his talk, I screwed up my courage, literally ran up to the podium, and I begged him to let me be his postdoc. A few phone conversations later, he said yes. Enthusiasm and persistence paid off.
Dig Deeper 3: How was the term “quorum sensing” coined?
Steve Winans is a quorum-sensing researcher at Cornell University. A relative of Steve’s coined the phrase "quorum sensing" at a Thanksgiving dinner. The relative is a lawyer named Rob Johnston. The account, as told by Steve Winans goes as follows: Steve Winans, "asked members of his extended family to help find a lively new phrase to describe the autoinducer-type pheromones. It was agreed that the terms "communulin" and "gridlockin" did not resonate. At some point between turkey and pie, a nonscientist in the group, Rob Johnston, mused that a high cell density could be viewed as a bacterial quorum. He then exclaimed, "Got it! Quormone!" In hindsight, it seems that the venerable terms "pheromone" and "autoinducer" suffice after all but that "quorum" appears to have answered some etymological need within the microbiology community." (Dunny, G.M. and Winans, S.C., 1999). Winans and colleagues later went on to use the term quorum sensing in a widely read early review of the process (1994) and the term stuck!
Dig Deeper 4: Northern Blot Analysis
A Northern blot was used to analyze the expression of the vqmR gene in V. cholerae. A gene is transcribed by RNA polymerase to make an RNA. Therefore, by analyzing the levels of the mRNA, one can assess whether the gene is turned on. In this method, the RNA is extracted from the bacteria. It is then pipetted into a well of an agarose gel and an electric current is applied (positive electrode at the bottom). The negatively charged RNAs migrate through the gel from top to bottom and become separated based on their sizes; smaller RNAs can move through the agarose gel faster and they end up closer to the bottom than larger RNAs. See Video DD 4 to better understand how a gel is run and how it can be used to analyze experiments. You can also run a simulated gel yourself in the Electrophoresis Activity by Douglas Koshland.
A Northern blot was used to analyze the expression of the vqmR gene in V. cholerae. A gene is transcribed by RNA polymerase to make an RNA, which then is translated by ribosomes into a functional protein. Therefore, by analyzing the levels of the mRNA, one can assess whether the gene is turned on and able to make a protein.
After running the gel, filter paper is laid on the agarose and the RNAs will be transferred from the agarose to the filter paper in their same relative locations (Figure DD 4). The filter paper is then incubated with a radioactive DNA that is complementary in its sequence to a specific RNA, in this case, the vqmR RNA. Because of this complementary, the radioactive DNA will hybridize (form a DNA-RNA double helix) with the vqmR RNA at its migration position on the filter paper. The filter paper is next washed to remove unhybridized radioactive DNA, and the washed filter paper is exposed to an X-ray film. The radioactivity from the DNA-RNA hybrid will appear as a dark band on the X-ray film when it is developed. From the position of the band, one can assess the size of the RNA and from its intensity one can infer the amount of RNA that hybridized with the DNA, which coincides with how much of that RNA was made by the cell.
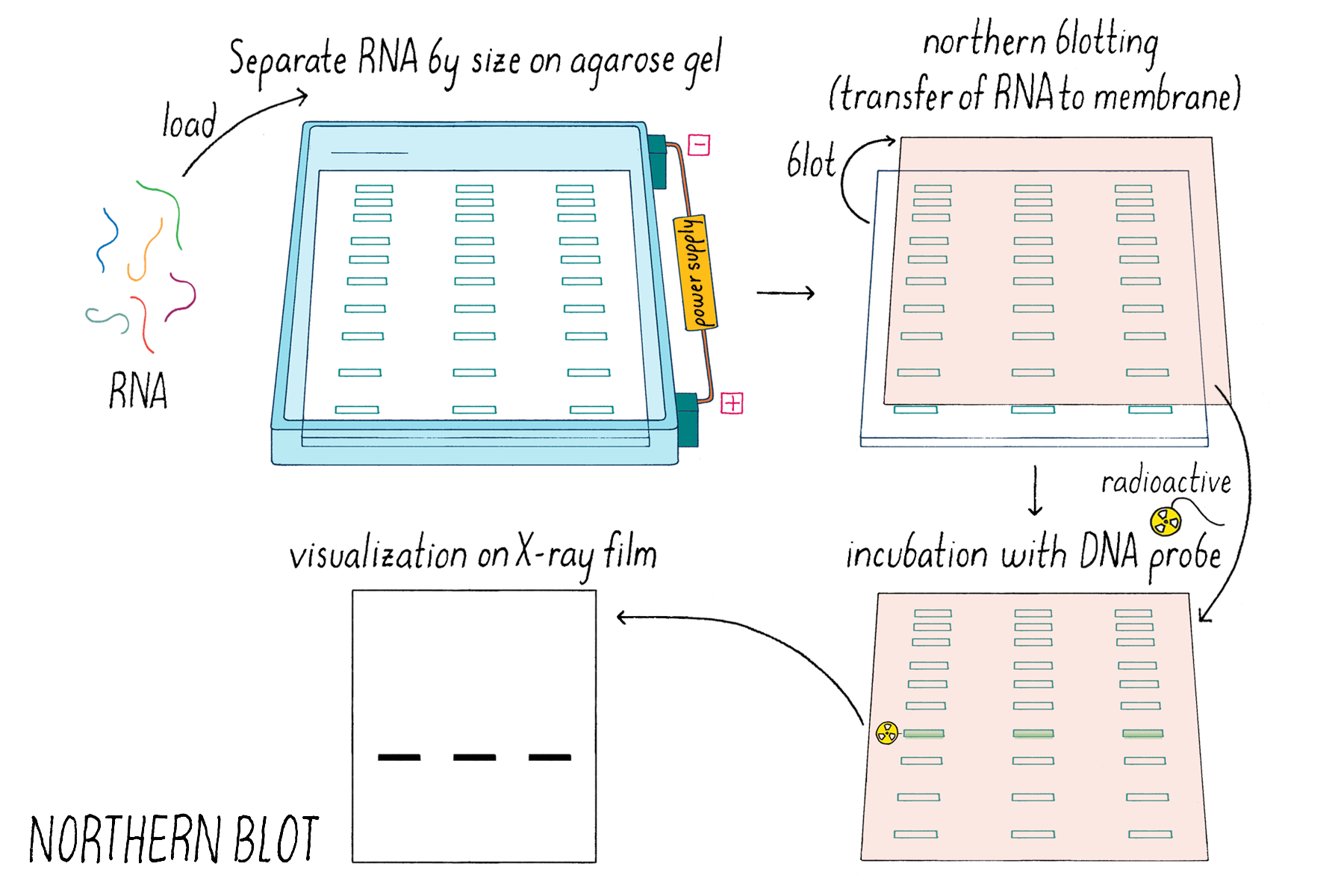
References and Resources
Guided Paper
Papenfort, K., Silpe, J.E., Schramma, K.R., Cong, J.P., Seyedsayamdost, M.R., and Bassler, B.L. (2017). Nat. Chem. Biol. 13:551-557.
This paper describes the discovery of a new quorum-sensing mechanism (and a new autoinducer called DPO) in the pathogenic bacterium Vibrio cholerae, as discussed in the Frontiers section of this Narrative.
DownloadReferences
- Tomasz A. Control of the competent state in Pneumonococcus by a hormone-like cell product: An example for a new type of regulatory mechanism in bacteria. Nature 1965;208:155–159.
This seminal paper shows that a substance released by bacterial cells can change the behavior of other bacterial cells that come in contact with it, suggesting that bacteria can coordinate behavior of the group. This work is highlighted in Clue 1 and Figure 3 in the Journey to Discovery.
- Nealson, K.H., Platt, T., and J.W. Hastings. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970;104:313–322.
A classic paper that established that bacterial bioluminescence is regulated by cell density. This work is featured in Clue 2 and Figure 5 in the Journey to Discovery.
- Eberhard, A. A.L. Burlingame, C. Eberhard, G.L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. Structural identification of an autoinducer of Photobacterium fischeri luciferase. Biochemistry 1981;20:2444–2449.
This manuscript reports the identification of the first bacterial autoinducer. See Clue 2 in Journey to Discovery.
- Engebrecht, J. K. Nealson, and M. Silverman. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 1983;32:773–781.
This landmark manuscript identifies the genes for bioluminescence and its regulation –the first quorum-sensing system. This paper has the experiment depicted in Figure 5 and the model shown in Figure 7. See Discovery Part A.
- Bassler, B.L., M. Wright, R.E. Showalter, and M.R. Silverman. Intercellular signaling in Vibrio harveyi: Sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 1993;9:773–786
- Bassler, B.L., M. Wright, M.R. Silverman. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence of function of genes encoding a second sensory pathway. Mol. Microbiol. 1994;13:273–286.
These two manuscripts report the discovery of quorum-sensing components in V. harveyi. The papers report that the V. harveyi components are not similar to those of V. fischeri, showing that quorum-sensing systems exist in bacteria other than V. fischeri and that very different signal transduction pathways can make up quorum-sensing systems. Moreover, this is the first work to demonstrate that there are multiple autoinducers involved. So, bacteria are multilingual! See Discovery Part B and Figure 10 and also Intra-species, Intra-genera, and Inter-species Communication and Figure 19 in Knowledge Overview.
- Dong, Y.-H., L.-H. Wang, J.-L. Xu, H.-B. Zhang, X.-F. Zhang, and L.-H. Zhang. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001;411:813–817.
This report describes the strategy to use the AiiA enzyme to degrade a pathogenic bacterium’s autoinducer rendering the bacterium incapable of causing infection. See Frontiers, Figure 20.
- Papenfort, K., J.E. Silpe, K.R. Schramma, J.P. Cong, M.R. Seyedsayamdost, and B.L. Bassler. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol. 2017;13:551–557.
This paper reports DPO a new quorum-sensing autoinducer involved in interactions between the microbiome and a bacterial pathogen. See Frontiers.
Resources
- Woody Hastings: Autoinduction: The discovery of quorum sensing. iBiology talk (11 min). Available at:
https://www.ibiology.org/speakers/woody-hastings/Woody Hastings describes his discovery described in Clue 2 in the Journey to Discovery.
- Margaret McFall-Ngai: The Hawaiian bobtail squid—Vibrio fischeri association. iBiology talk (28 min). Available at:
https://www.ibiology.org/microbiology/vibrio-fischeri/This video describes the symbiotic relationship between a squid and bioluminescent bacteria.
- Bacterial Quorum Sensing Experiment Slides. Available at: http://www.hhmi.org/biointeractive/bacterial-quorum-sensing
Dive deeper into quorum sensing by working through experiments concerning genetically-engineered bioluminescent bacteria.
- Q&A on Quorum Sensing. Available at: http://www.hhmi.org/biointeractive/qa-quorum-sensing
In this video, Dr. Bonnie Bassler answers questions regarding quorum sensing and other related topics.
- The Molecular Cascade in Bacterial Quorum Sensing. Available at:
http://www.hhmi.org/biointeractive/molecular-cascade-bacterial-quorum-sensing.
This video describes the molecular processes that are at the center of bacterial quorum sensing.
- The Lux Operon Controls Light Production. Available at: https://www.hhmi.org/biointeractive/lux-operon-controls-light-production
A short animation that describes how the lux operon works to cause bioluminescence in bacteria.
- TED Talk with Bonnie Bassler Describing How Bacteria Communicate. Available at: https://www.ted.com/talks/bonnie_bassler_on_how_bacteria_communicate
In this TED talk, Dr. Bonnie Bassler discusses the ins/outs and implications of bacterial communication.
- NPR: A Virus Can Eavesdrop On Bacterial Communication, by Susan Brink. Available at: https://www.npr.org/sections/goatsandsoda/2018/12/13/676389858/a-virus-can-eavesdrop-on-bacterial-communication
- PBS’s NOVA: Chatty Bacteria Might Be Most Vulnerable to Viruses, by Katherine Wu. Available at:
https://www.pbs.org/wgbh/nova/article/chatty-bacteria-might-be-most-vulnerable-viruses/New stories about how viruses can eavesdrop on bacteria.