Lighting up Biology
Expression of the Green Fluorescence Protein
Martin Chalfie Ghia Euskirchen
 Martin (Marty) Chalfie
Martin (Marty) Chalfie
Martin (Marty) Chalfie holds the title of University Professor at Columbia University in New York City. He shared the 2008 Nobel Prize in Chemistry, along with Osamu Shimomura and Roger Y. Tsien, "for the discovery and development of the green fluorescent protein, GFP." He received his Ph.D. in Physiology from Harvard University in 1977.
 Ghia Euskirchen
Ghia Euskirchen
Ghia Euskirchen received a Ph.D. in molecular biology from Columbia University. Her career has taken her to Yale University as a postdoc, then to Stanford University where she was director of a DNA sequencing center. She currently resides in Washington, DC.
What’s the Big Deal?
Certain jellyfish emit a beautiful green light. Although why jellyfish would want to glow in the dark is still a mystery, the green fluorescent protein (GFP) from these animals has proven to be a gift from nature that has transformed cell biology. Three scientists were recognized with a Nobel Prize for their work on GFP, although others contributed in important ways, as you will see from this story. Why is GFP a big deal? With the combination of GFP and microscopes equipped with very sensitive cameras, scientists began making incredible movies of the dynamic processes that underlie life at that microscopic level. Perhaps you have seen “Planet Earth” and the amazing cinematography of animals in these videos. Now, equally amazing movies are being made of the inner workings of living cells, allowing us to see when and where genes are expressed and other dynamic processes that enable life. GFP has given us the tools to make “Planet Cell.” GFP also unleashed the amazing creativity of scientists. GFP has been engineered into “endless forms most beautiful and most wonderful” (to quote Darwin). There are now fluorescent proteins of all colors, variants that can be switched on and off, and modified versions that sense and report upon conditions inside of cells. And it all started with a deceptively simple experiment. The story of GFP also contains a lesson for young scientists—don’t be dissuaded by other people who might say “no, that experiment won’t work.” We will let Marty Chalfie and Ghia Euskirchen take you on a tour of what they did, giving you a front row seat to the birth of the GFP revolution.
Learning Overview
Big Concepts
The “green fluorescent protein” (GFP) has revolutionized cell biology because it provides a fluorescent beacon that enables scientists to follow individual molecules in a living cells or the behavior of whole cells in living organisms. This key experiment (first done by a new graduate student) also provides a window into the process of gene cloning, how GFP is being used in biology, and into the serendipitous nature of science itself.
Bio-Dictionary Terms Used
Amino acids, antibody, bioluminescence, Central Dogma, DNA, enzyme, fluorescence, gene, mRNA, mutant, neuron, plasma membrane, protein, Phosphorylation
Terms and Concepts Explained
C. elegans, DNA cloning, E. coli, Fluorescence, immunofluorescence, mitotic spindle, polymerase chain reaction, reporter, transformation, restriction enzyme
Introduction
-
Certain jellyfish (including Aequorea victoria) possess a protein that can absorb blue light and emit green light. This protein is called the green fluorescent protein or GFP.
What Events Preceded the Experiment?
-
Before GFP, scientists used a technique called “immunofluorescence” to reveal the location of a protein within a cell. However, this method involved chemical fixation, which killed the sample a and precluded observation of cellular dynamics. If GFP could be expressed in living cells other than the jellyfish, then live cell observations could be made.
Setting up the Experiment
-
Martin Chalfie, his student Ghia Euskirchen, and collaborator Douglas Prasher undertook an experiment to express the cloned jellyfish GFP DNA in bacteria and see if the living bacteria would glow green.
-
Many people thought that this experiment would not work. GFP undergoes a chemical reaction that produces a ring within its original linear backbone, which is critical for its fluorescence. Several researchers at the time speculated that one or more jellyfish enzymes might be needed for this reaction. If this speculation were true, bacteria lacking these hypothetical enzymes could not fluoresce green.
Doing the Key Experiment
-
The scientists used polymerase chain reaction and cDNA cloning to introduce the jellyfish GFP gene on a plasmid into bacteria. Bacteria that successfully took up the plasmid were selected using an antibiotic resistance marker.
-
The scientists induced the expression of the GFP gene. A control experiment was performed in which bacteria were transformed with a plasmid that did not contain the GFP gene. They looked to see if the bacteria with the GFP gene became fluorescent whereas the control bacteria did not.
-
The experiment worked on the first try. The bacteria with the gene, but not the control, became fluorescent. No additional enzymes from the jellyfish were needed.
-
GFP then was used as a “transcriptional reporter” to analyze what genes are turned on in particular cells. The GFP gene also was joined onto other genes to create “fusion proteins”, where the GFP fluorescence could be used to track the whereabouts of proteins in cells.
What Happened Next?
-
New fluorescent proteins with different colors were discovered or made. Numerous uses of GFP evolved in biotechnology and cell biology. Because of the importance of GFP as a tool, three scientists were awarded the Nobel Prizes for discoveries related to GFP.
Closing Thoughts
-
The GFP story reveals how important new technologies can emerge from studying unusual organisms (jellyfish), why persistence pays off, and how serendipity takes one down unexpected paths of discovery.
Guided Paper
M. Chalfie, Y. Tu, G. Euskirchen, W.W. Ward, and D.C. Prasher. Green fluorescent protein as a marker for gene expression. Science 1994;263:802–805.
This paper describes the discovery of green fluorescent protein (GFP), which allows scientists to monitor gene expression and protein localization in living cells. This research led to Chalfie receiving his Nobel prize in 2008.
DownloadIntroduction
I am famous in my department for sleeping through scientific talks, but on April 25, 1989, I heard one that kept me wide awake. I am a faculty member at Columbia University, and for many years, those of us interested in neuroscience met every Tuesday at noon to hear a scientific seminar. That Tuesday, the speaker, Paul Brehm, grabbed my attention. He began by describing the work of Osamu Shimomura on bioluminescence, the ability of different organisms to generate light.
I had known of one of Shimomura’s discoveries—the isolation of the bioluminescent protein aequorin from the jellyfish Aequorea victoria. Aequorin produces a bright blue light in the presence of calcium. What I hadn’t known until Paul’s seminar was that Shimomura had discovered a second protein, a protein that he initially called the Green Protein and that we now call the Green Fluorescent Protein or GFP. GFP was going to change my life.
Osamu Shimomura’s discovery of these two proteins are themselves a study in overcoming scientific frustration. He started out with a fascinating problem: how do different organisms generate light? Many organisms (e.g., fireflies, glow worms, bacteria, fungi, fish) generate light, but they do so in many different ways (see also, the story of bioluminescent bacteria in the Narrative on Quorum Sensing by Bassler). Shimomura wanted to find the molecules that allowed the jellyfish to produce a beautiful and intense green light. However, isolating the light-producing protein from the jellyfish proved elusive.
One night, after another frustrating day of failure, he decided to clean up and go home for dinner. He threw all of his seemingly worthless protein samples into a sink that also had an overflow from seawater tanks. Shimomura turned off the lights and was about to walk out the door, when he looked back at the sink. It was glowing! Something from his protein samples was generating light. As you can imagine, literally seeing the light made him stop and think about what was happening. After some thought, he realized that it must have been the seawater that mixed in with his protein and caused light to be produced. Seawater contains calcium, something he had never tried adding in his experiments. Perhaps calcium was needed to trigger a jellyfish protein to make light? A few days later he showed that his guess was correct; addition of calcium to his preparation produced a flash of light. He then used this ability to stimulate light production to guide him in the purification of the aequorin protein.
Unfortunately, Shimomura’s discovery of aequorin left him with another problem: the light emitted by aequorin was blue instead of the green glow emitted by the jellyfish. Some other factor (another protein perhaps?) must have converted the blue light to green. When he tested other jellyfish protein samples, he indeed found another protein that could absorb blue light and re-emit it as green light. That protein we now call GFP.
What events preceded the experiment?
When I heard about GFP in that seminar in 1989, I was so excited that I spent the rest of the seminar time thinking about the experiments I wanted to do and was not paying attention to the speaker. However, I didn’t fall asleep. To understand why I was so excited to hear about Shimomura’s discovery of GFP, you need a bit of background about what we were doing in the lab at the time.
Starting in 1977, I began using the small (1 mm) nematode (round worm) called Caenorhabditis elegans (C. elegans, for short) to identify and study genes involved in mechanical sensation (several of our senses, hearing, touch, detection of blood pressure involve detecting a mechanical perturbation). Once we found a potentially interesting gene through genetic studies, one of the first things that we wanted to know was what cells in the worm were expressing the gene. When a gene is expressed (“turned on”) in a cell, then an mRNA is transcribed, and a protein is made from the mRNA (see Central Dogma). We were particularly interested in genes that were expressed in the neurons that sensed a gentle touch stimulus.
At the time, scientists typically looked to see whether a cell turned on a gene to express a protein using a method called “immunofluorescence” (Figure 1). For this method, you first need to make an antibody against the protein of interest. Then, the worm (or other animals/cells) is “fixed” with chemicals that preserve cell structure, but which kills the animal in the process. The fixed cells are then incubated with the antibody that binds to your protein of interest and but not to other proteins in the cell. However, the antibody needs to be made visible with a fluorescent dye that emits light so that one can see it under a microscope. Frequently, a second commercially sold antibody labeled with a fluorescent dye is used to bind to the first antibody. The fluorescent signal pinpoints the location of your protein of interest. In the example shown in Figure 1, the protein of interest is localized to the plasma membrane.
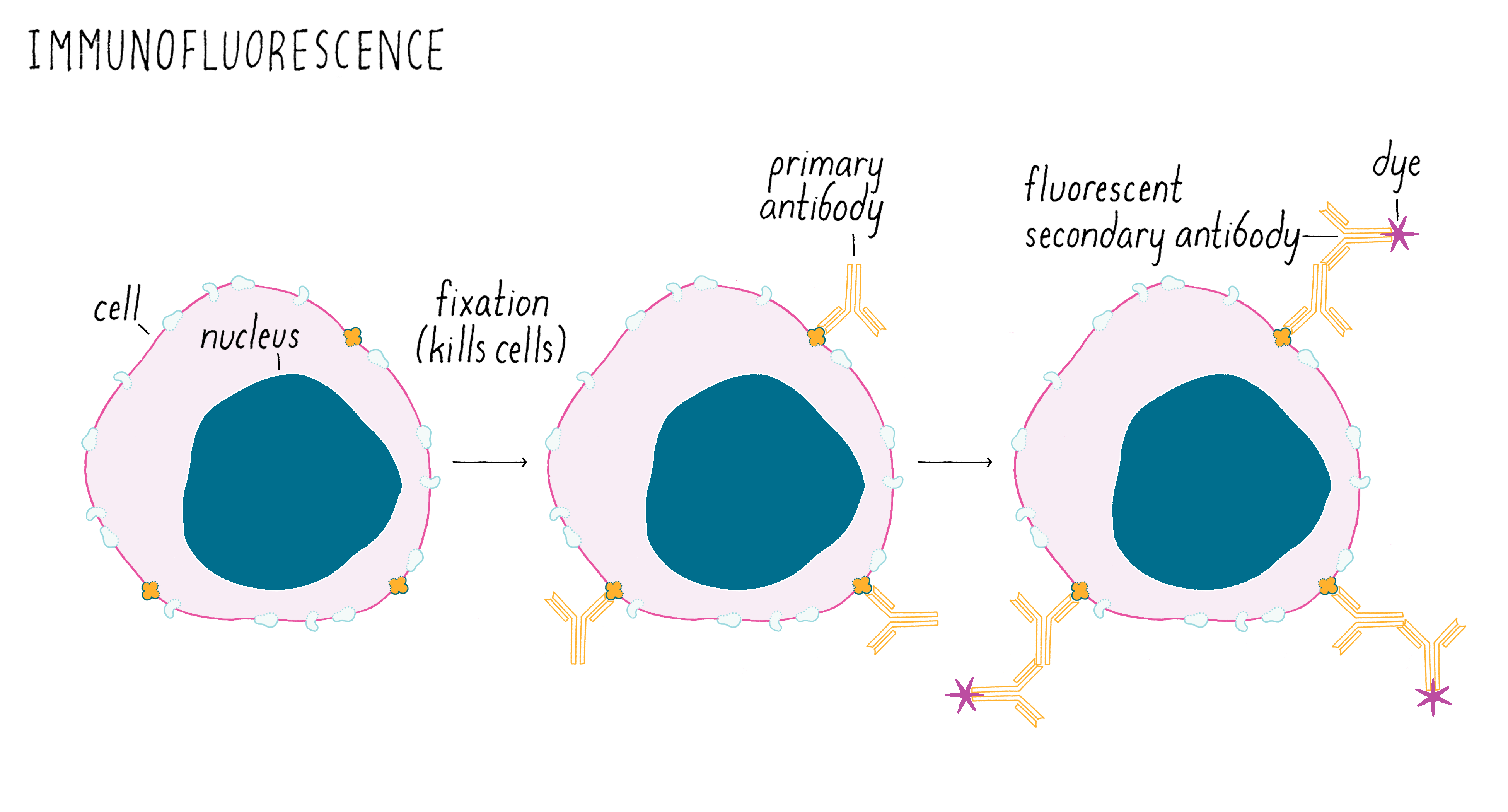
Figure 2 shows a real immunofluorescence image of a protein localized to the plasma membrane of a human white blood cell. The image is taken with a sensitive digital camera attached to a fluorescence microscope. The purple fluorescent antibody bound to the protein reveals a rim outlining the perimeter of the cell, which is the plasma membrane. The nucleus in the center is stained with a fluorescent dye that binds to DNA.
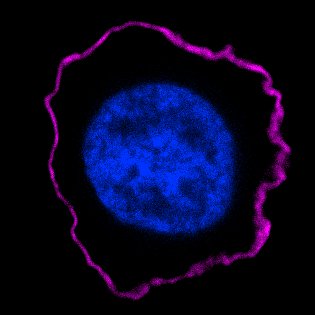
The main problem with immunofluorescence is this procedure kills the cell; once dead, one cannot follow changes over time. Immunofluorescence provides a snapshot but not a movie.
My mind was racing during the seminar. What if GFP could be used as a reporter for whether a particular gene was expressed in a live animal? Then we could shine blue light on a living animal and see which cells lit up as green. And we could see how those cells changed over time, such as during development, which was my interest. I have often wondered why no one else in the room seemed as excited as I was, but it may have been because I was the only one trying to find out where genes were expressed in a transparent animal (yes, you can shine light through the C. elegans worm). Because of its transparency, one should be able to see the fluorescent light emitted by GFP in C. elegans. GFP could serve as a lantern illuminating where a particular cell type is located within an organism and what it is doing without needing to kill the animal.
No wonder I was too excited to listen to the seminar.
Setting up the experiment
The next day I started learning more about GFP, mainly by phoning people. One encouraging fact was that GFP did not need anything added to be fluorescent. i.e., to absorb blue light and emit green light. What I was really hoping to uncover in all my phone calls was a person who was working on obtaining the region of the jellyfish DNA that contained the gene that encoded GFP. We needed the GFP DNA so that we could transfer it and express it in C. elegans. Luckily, I found someone working to identify the GFP gene, Douglas Prasher, a researcher at the Woods Hole Oceanographic Institute. When I reached Douglas by phone, I found that he had very similar ideas about using GFP and at the end of our hour-long conversation, we decided to collaborate.
Douglas said that he would call when he had succeeded in the first step—isolating the jellyfish DNA for GFP. But at that time, this step was not easy. If you want to know more about how he did this, see the “Dig Deeper 1: cloning jellyfish DNA.”
Unfortunately, I never got Douglas’ call that he succeeded in cloning GFP (I later learned that he called me when I was away on sabbatical). I subsequently learned that because Doug couldn’t get in touch with me, he thought that I had left science.
In September 1992, the new graduate students joined the department and one of them, Ghia Euskirchen, asked if she could rotate in my lab (many graduate students do short-term projects called “rotations” to try out several labs before deciding where to do their graduate research). I was very happy to have her rotate in my lab, because she had already obtained a master’s degree in chemical engineering at Columbia working on fluorescence. I told Ghia about my idea of using a fluorescent protein as a biological marker. But after waiting 3 years for Douglas’ call, I had given up hope about his experiments. So I suggested that we might have to look for something other than GFP. We then looked up the term “fluorescent protein” on Medline (an early search engine made by the National Institutes of Health) and see what other fluorescent proteins existed. Columbia had just connected the University computers to Medline, another fortunate bit of timing. The first paper that popped up in our search was Douglas’ paper from earlier that year describing the isolation of the GFP DNA. We ran down to the library to find the paper. Then, another stroke of luck: although Douglas had left the Woods Hole Oceanographic Institute, the paper had his new phone number (scientific articles do not often list authors’ telephone numbers). I immediately called Douglas and renewed our collaboration. He sent us the DNA clone a few days later.
With the jellyfish GFP DNA in hand, Ghia’s project was, in one sense, very straightforward: introduce the jellyfish GFP DNA into bacteria (E. coli), express the protein, and see whether the bacteria emitted green light when stimulated by blue light.
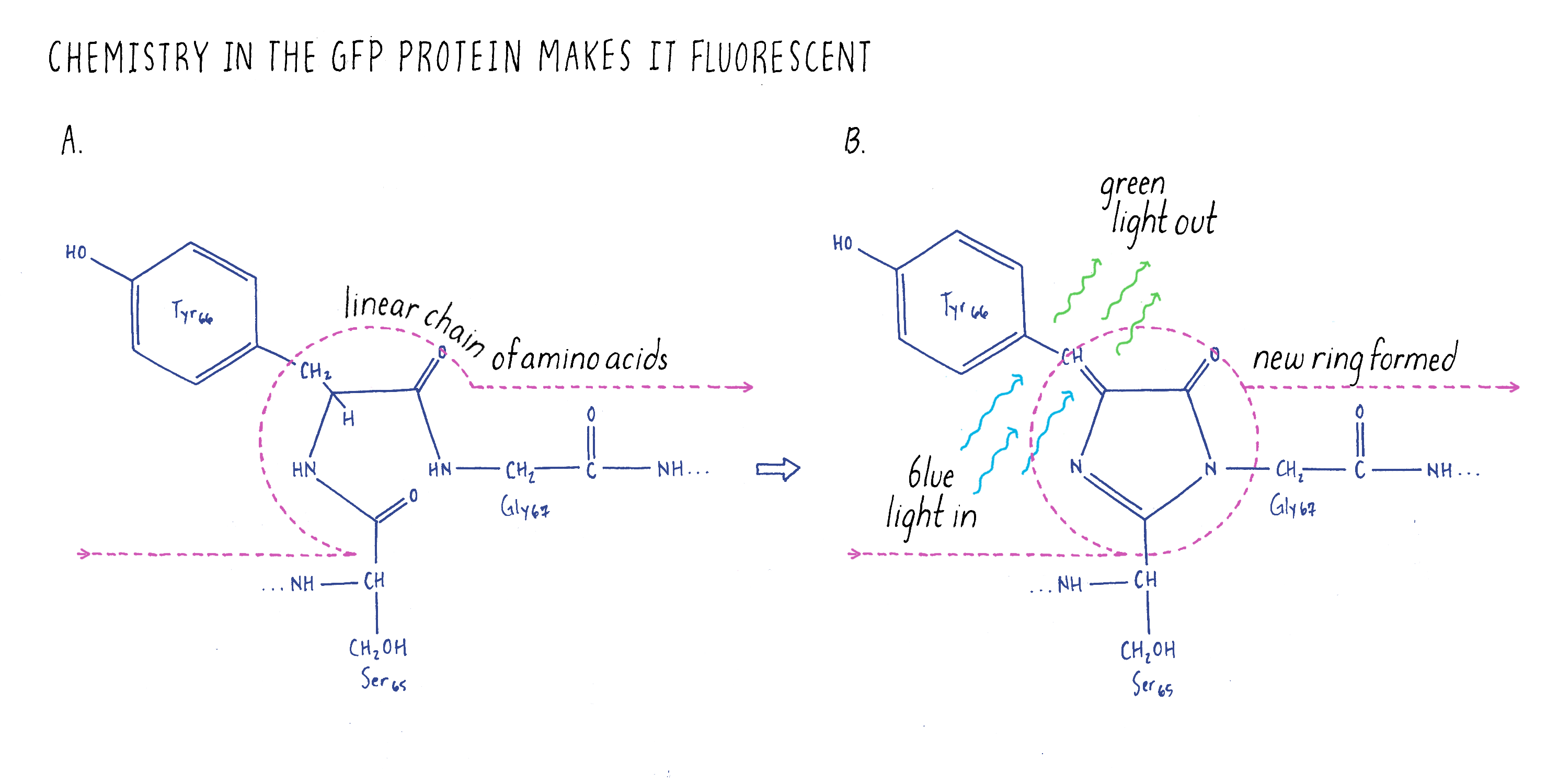
Most people working on GFP at the time believed that this experiment would never work because GFP is a very unusual protein. Most proteins are linear strings of amino acids. GFP also starts out with the usual linear peptide backbone (the line shown in Figure 3), but then it undergoes a chemical rearrangement to make a loop (a chemical ring) in a section of the linear backbone (Figure 3). This chemical ring is essential for the protein to be fluorescent. When we did our experiments, no one knew how this loop was made. People imagined that there was a jellyfish-specific enzyme that produced the loop. Thus, if this enzyme was not present in bacteria (as one would expect), then GFP should not be fluorescent. As a result, our experiment would be considered a fool’s errand; the bacteria should not glow green. However, we were not dissuaded and decided to try the experiment anyway.
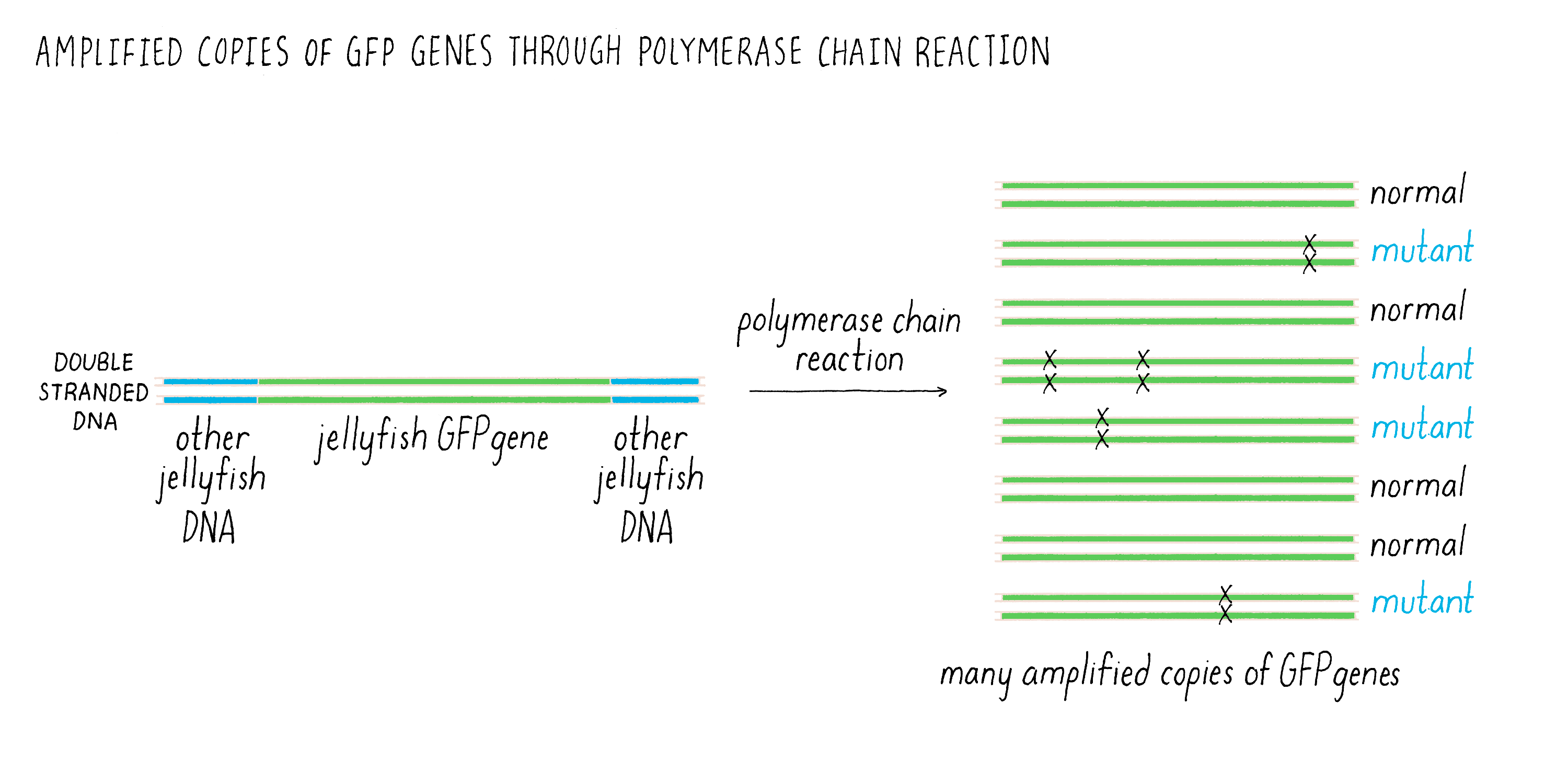
There was one additional experimental detail, which proved to be very important and proved fortunate for us. Douglas’ GFP clone had the complete coding sequence for GFP but it also had additional jellyfish DNA on either side. This adjacent DNA did not code for amino acids and we knew nothing about its function. I was leery of this extra DNA and did not want to include it in our experiment and suggested to Ghia that we leave it out and just use the DNA that coded for GFP. We could copy just the GFP coding sequence using the polymerase chain reaction (or commonly known as PCR), a technique that amplifies a precise piece of DNA. See Video 2 for an explanation of how PCR works.
The one problem with PCR at this stage of its development is that it was somewhat error prone (i.e., copying the DNA by PCR could introduce mutations (Figure 4). Thus, researchers who wanted accurate copies of the GFP DNA would have left the jellyfish DNA alone and used the entire DNA fragment obtained by Douglas to express the GFP. In contrast, I reasoned that if some of the PCR DNA products had mutations, this would not be a problem. Certainly some of the DNA would have been copied accurately by PCR, and those bacteria would be easy to spot because they would be glowing green. As you will read later, this seemingly trivial strategic decision allowed us to succeed, while others’ laboratories failed in the same effort using Douglas’ original DNA clone.
Now I will turn the narrative over to Ghia who can tell you what she did.
Doing the key experiment
Ghia
Molecular biology methods were completely new to me when I was handed the GFP DNA clone. As a first year Ph.D. student, I was anxious about proving myself in a both new discipline and in a new department. I was confident in my fluorescence microscopy and bacteriology skills, but I had never previously set up a PCR reaction. Although it’s difficult to imagine, many resources we now take for granted either didn’t exist or weren’t prevalent in 1992. Journal articles had to be photocopied from large bound volumes during library hours (pdfs didn’t yet exist), photomicroscopy required film (again no digital option), and without graphical web browsers, new methods propagated from lab to lab on a time scale of years instead of days.
Bench work is a balance between optimism and contingency planning. Although we were hopeful that E. coli expressing the jellyfish GFP gene would exhibit some sort of encouraging signal of emitted light, conventional wisdom held that GFP probably would not fluoresce on its own without a jellyfish helper protein. If so, we would then have to hunt for this helper protein. As my lab notes stated, “Apoprotein (protein before the fluorophore is made) probably will not fluoresce. Clone modification enzyme.”
Nonetheless, I set forth with a strategy that Marty outlined during our initial discussions. We would copy just the protein coding region of the jellyfish DNA using PCR. PCR generates millions of copies of GFP DNA in a test tube (see Video 1) that could then be cloned into a plasmid suitable for expressing the protein in E. coli. Once the PCR reaction was completed, the next step was to insert the PCR fragment into the bacterial plasmid (Figure 5). This was done by cutting the DNA plasmid at a specific site using a restriction enzyme that would generate single-stranded DNA that overhang at the ends. These are called “sticky” ends were because they could interact with complementary bases from sticky ends created by enzyme cutting of the PCR-generated DNA fragment. In this manner, the smaller PCR DNA could be inserted into a larger plasmid that is capable of replicating inside of bacteria. After sticking together, there are still breaks where the DNA backbone is not connected between the GFP fragment and plasmid. These cut sites need to be healed to make a circle of continuous DNA. Another enzyme is then added to connect (“ligate”) the PCR DNA and plasmid DNA into a continuous circle of double-stranded DNA. This overall process of moving a gene of interest into a plasmid that can replicate and make many copies of that gene is usually what scientists refer to a “cloning a gene.”
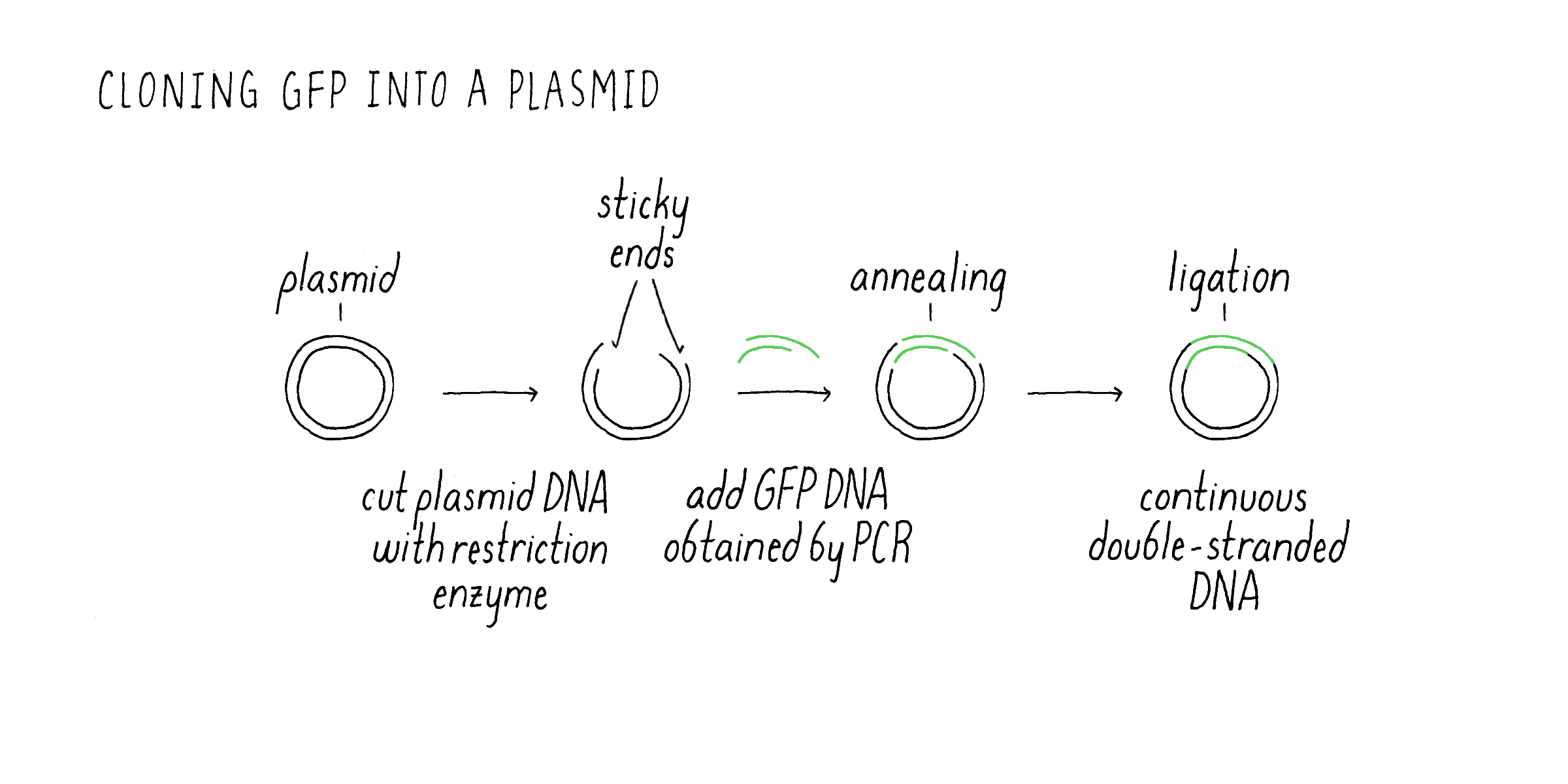
After preparing the GFP plasmid, the next step was getting inside of the bacteria E. coli. This uptake process is called “transformation” (Figure 6). Once inside of bacteria, the plasmid can replicate, i.e., it can make new copies of itself as the bacteria also divides. The plasmid also contained an additional gene that confers resistance to an antibiotic. E. coli that do not have the plasmid die in the presence of the antibiotic, while the transformed bacteria containing the GFP plasmid will live. In case that expressed GFP might be toxic to the bacteria, I used a plasmid that had what is called an inducible promoter. Normally, the promoter is “off” and GFP mRNA and protein are not made. However, upon addition of a special sugar called IPTG, the promoter is turned “on” and RNA is made and then translated into the GFP protein.
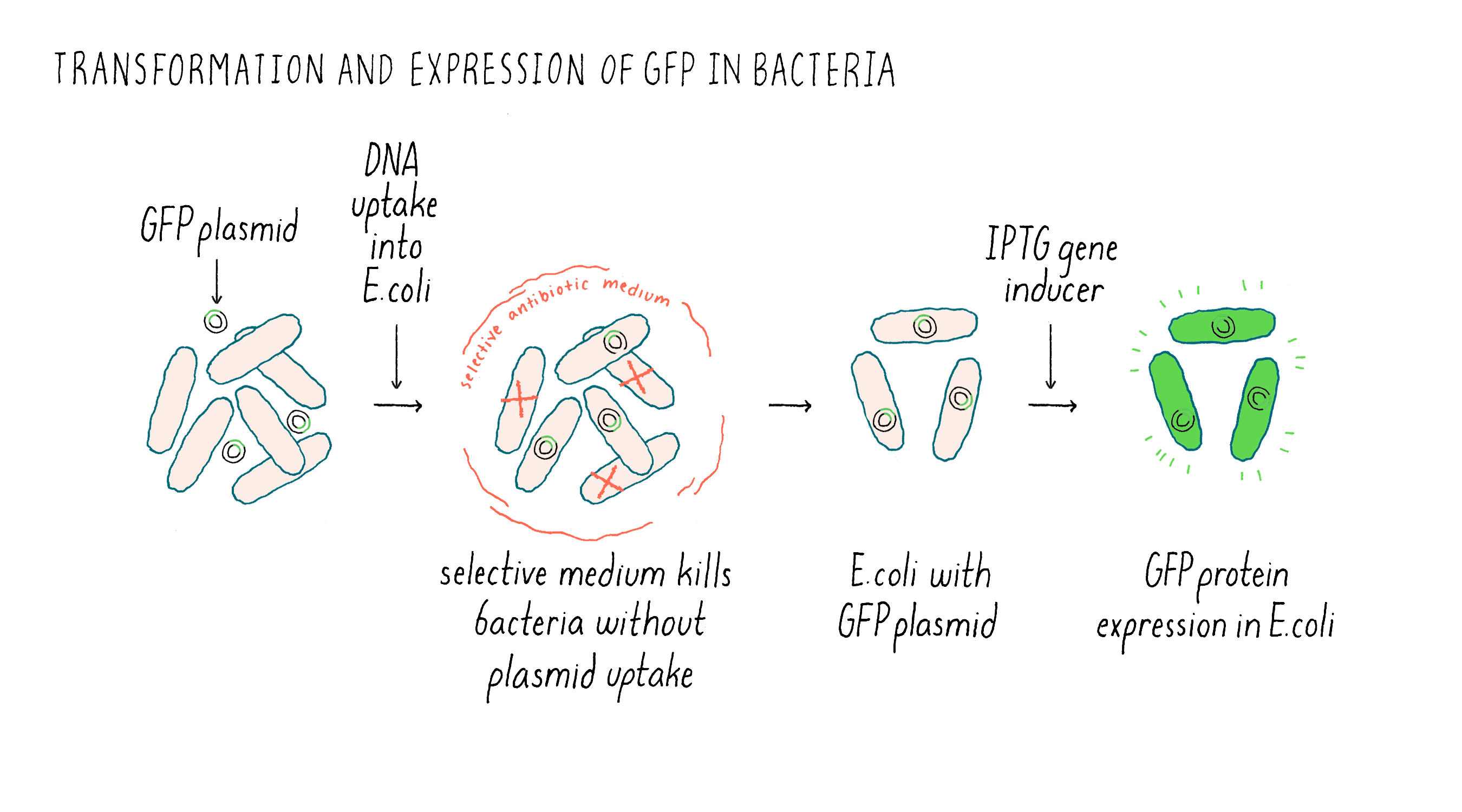
After selecting for E. coli with the GFP expression vector, I was finally ready for the moment of truth. As a first-year graduate student, coursework and experiments were intermixed with lectures during the weekdays and lab work during the evenings and on weekends. Consequently, it was a Tuesday evening when I added the inducer IPTG to turn on the expression of the GFP gene in my E. coli cultures. Two hours later, the cultures were ready to be examined for green fluorescence. This portion of the experiment required a microscope equipped with a special bulb and filter set to focus light with the right wavelength that would excite the GFP fluorophore. I had some idea of the microscope settings required based on publications that studied the biochemistry of GFP purified from jellyfish; however, some fiddling was likely because to my knowledge no one had previously expressed GFP in E. coli.
Unfortunately, when I peered into the microscope, the GFP-transformed E. coli were disappointingly dark. I wasn’t sure if this was due to the lack of GFP fluorescence, my own fumbling with a microscope that I hadn’t used before, or that the microscope hardware was not set up to detect the green wavelength of GFP. Fortunately, I still had a key to my former engineering lab with a familiar fluorescence microscope, which would allow me to test at least one of the possible reasons for failure. I gathered my experiment and headed to the engineering building. By now it was late, nearly midnight. I settled in with a familiar microscope in a darkened room for the best observation conditions. This time, however, I was stunned: the GFP-transformed E. coli were actually fluorescing green! Just to be certain, I compared the GFP-transformed E. coli to E. coli that had undergone the same protocol but had been transformed with a plasmid without the GFP coding sequence. This is known as a “control experiment” whose purpose was to make sure that the green image I was seeing was indeed due to the presence of GFP gene.
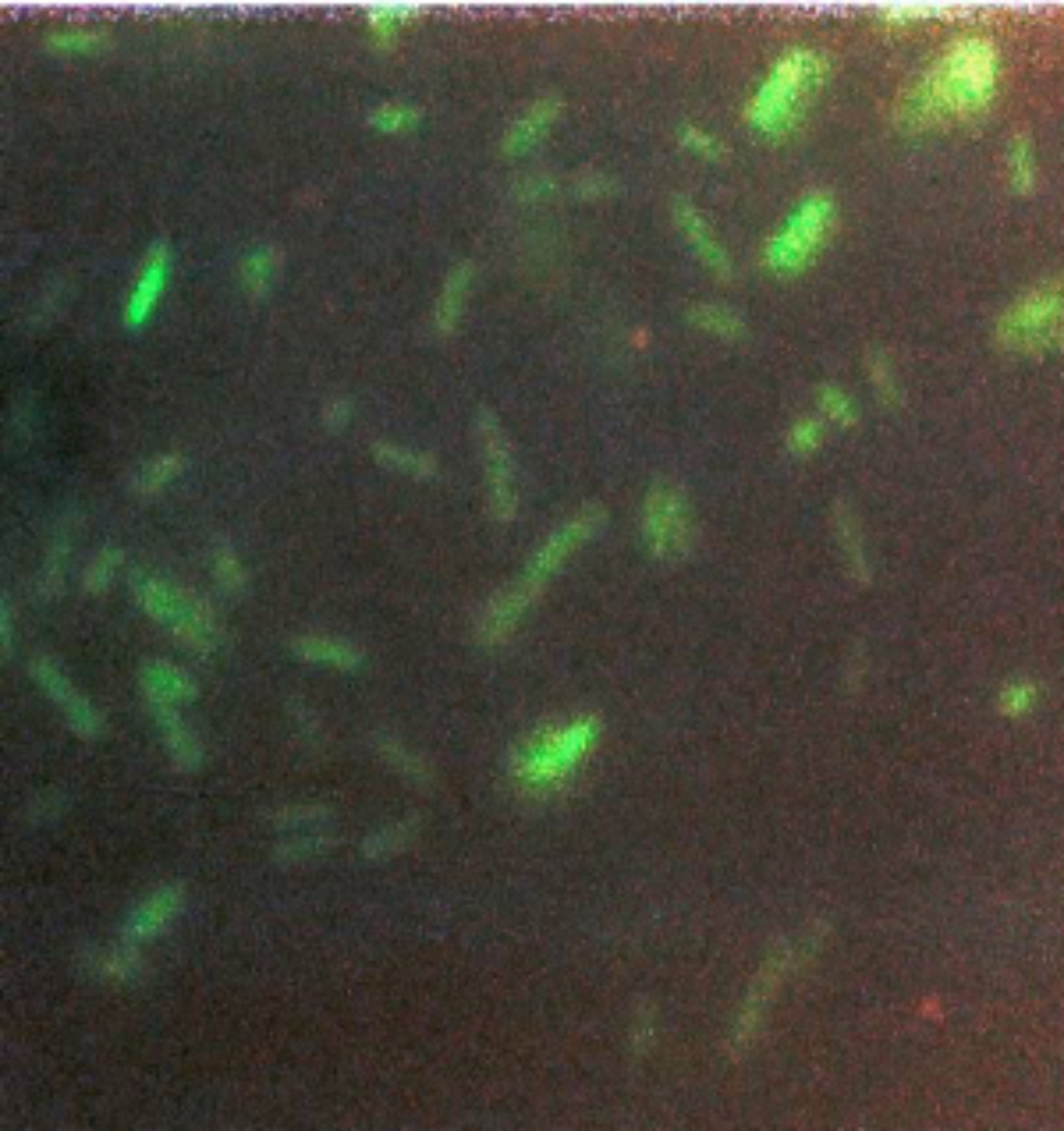
The E. coli transformed with the plasmid without GFP were dark and the GFP-transformed E. coli were green (Figure 7). This was the best-case scenario and one that nobody expected.
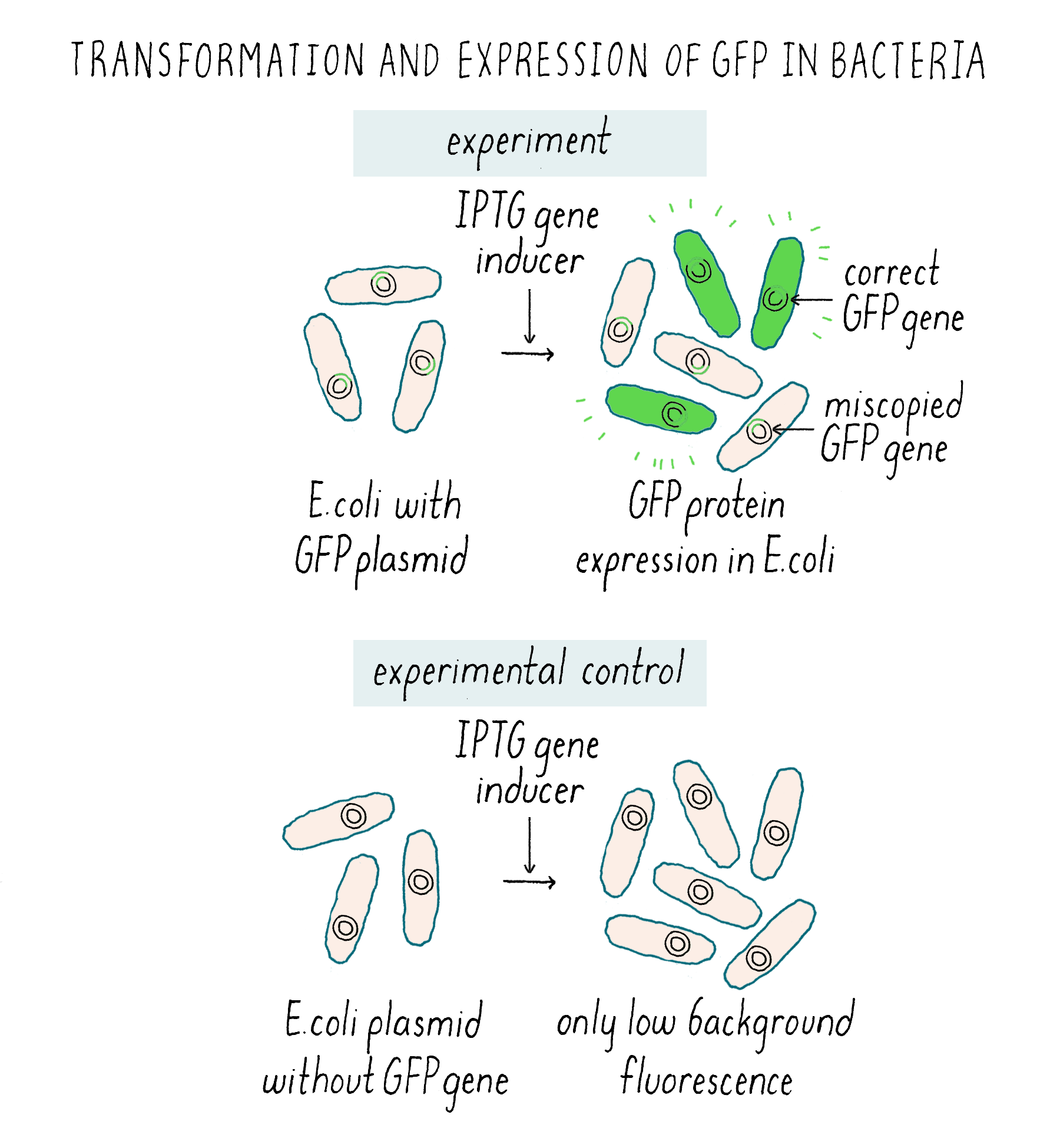
I knew it was a big moment (although just not how big) and I took some color photos that evening (Figure 8) to document the glowing green bacteria.
Still stunned, I sat in the dark for a few moments longer and collected my thoughts. It occurred to me that no one would believe this result especially as I barely believed it myself. Using pre-digital photography, I had no idea if the photos would come out until the film was developed, which wouldn’t be for at least a day. Trembling with excitement, I dashed back to the Chalfie lab to recruit witnesses. Needless to say, Marty was ecstatic when he heard and saw the results for himself.
Marty
Ghia’s result was great news; we were all very excited about the potential of GFP as a biological marker. This one experiment told us that GFP could be made in a living organism without introducing extra factors from the jellyfish. We now know that the chemistry that produces the unusual chemical ring needed for fluorescence (Figure 3) happens spontaneously after the GFP protein is synthesized; no extra enzymes are needed.
Unbeknownst to us, several other laboratories thought of using GFP as a biological marker (as often happens many people think of the same idea), but their experiments hadn’t worked. Again, we were fortunate. As it turned out, our choice to remove the extra jellyfish DNA on either side of the GFP gene turned out to be why our experiments worked and others failed. To this day, we still do not understand why those extra bits of jellyfish DNA prevent the production of GFP. Thus, by being careful in what we chose to include, but not in how accurately it was copied, our experiment ended up being successful.
One final comment about Ghia’s experiment: her successful use of the fluorescence microscope in her old lab in the Engineering School meant that we had a problem: we did not have our own microscope to do future experiments. We also soon wore out our welcome on other people’s microscopes. I solved this problem by calling up the sales representatives of various microscope companies and told them that we had just developed a new method to look at gene expression and were in the market for a new microscope. I then asked for loaner microscopes to test before buying. Amazingly, this ploy worked and we finished off the project using loaner microscopes.
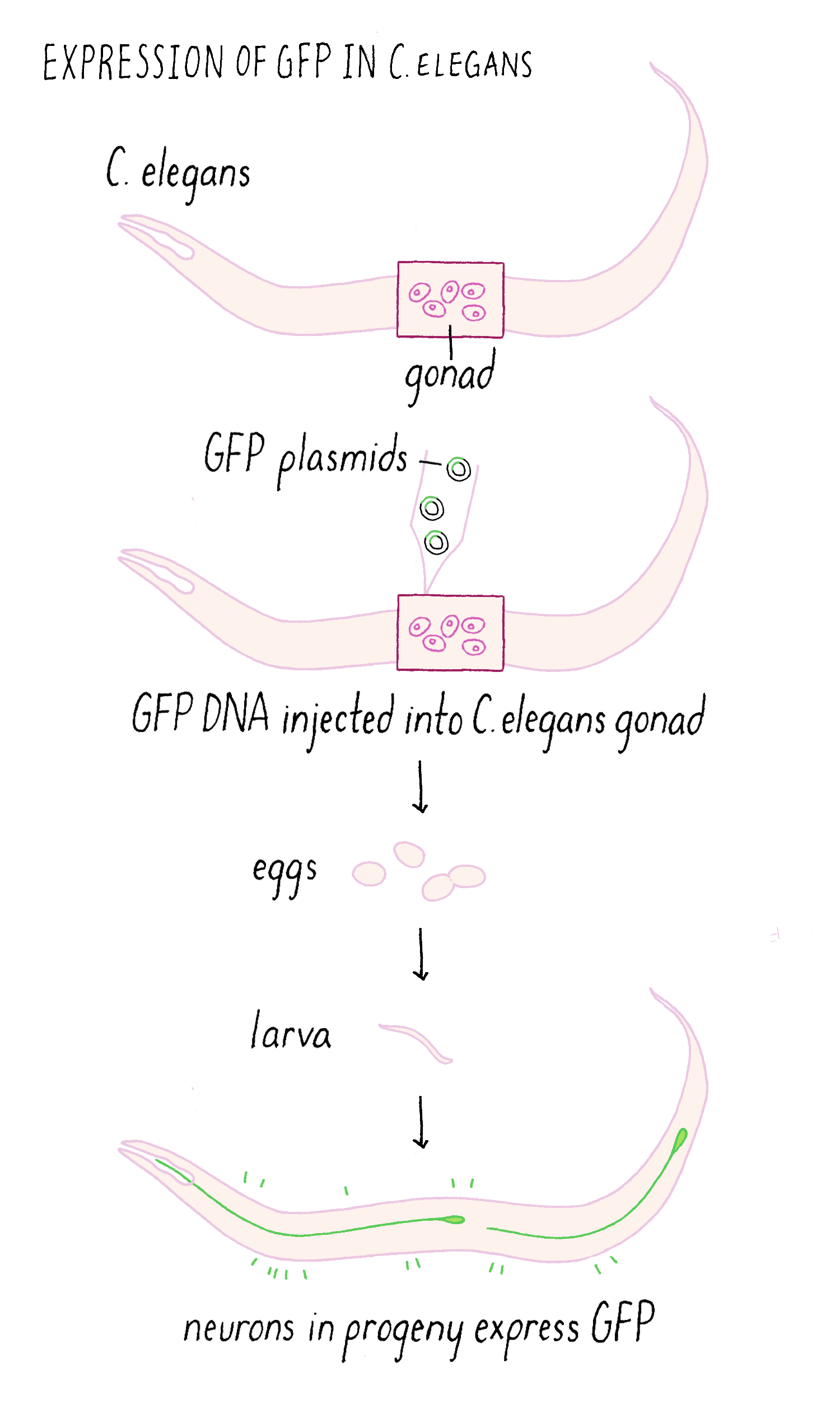
Making glowing bacteria was wonderful, but I wanted to return to my original motivation—expressing GFP in the worm C. elegans and be able to visualize individual cells in live animals. A talented technician, Yuan Tu, did this experiment and showed that GFP can be expressed in a functional fluorescent form in worm nerve cells, again without needing any other factors. To get the GFP to only express in only a certain population of neurons, we had to use an added trick of using a special promoter that would only turn on the expression of the GFP gene in touch neurons but not the other cells (Figure 9). With this “transcriptional reporter,” only the neurons that we wanted to study would make the GFP protein and be “lit up” with fluorescence.
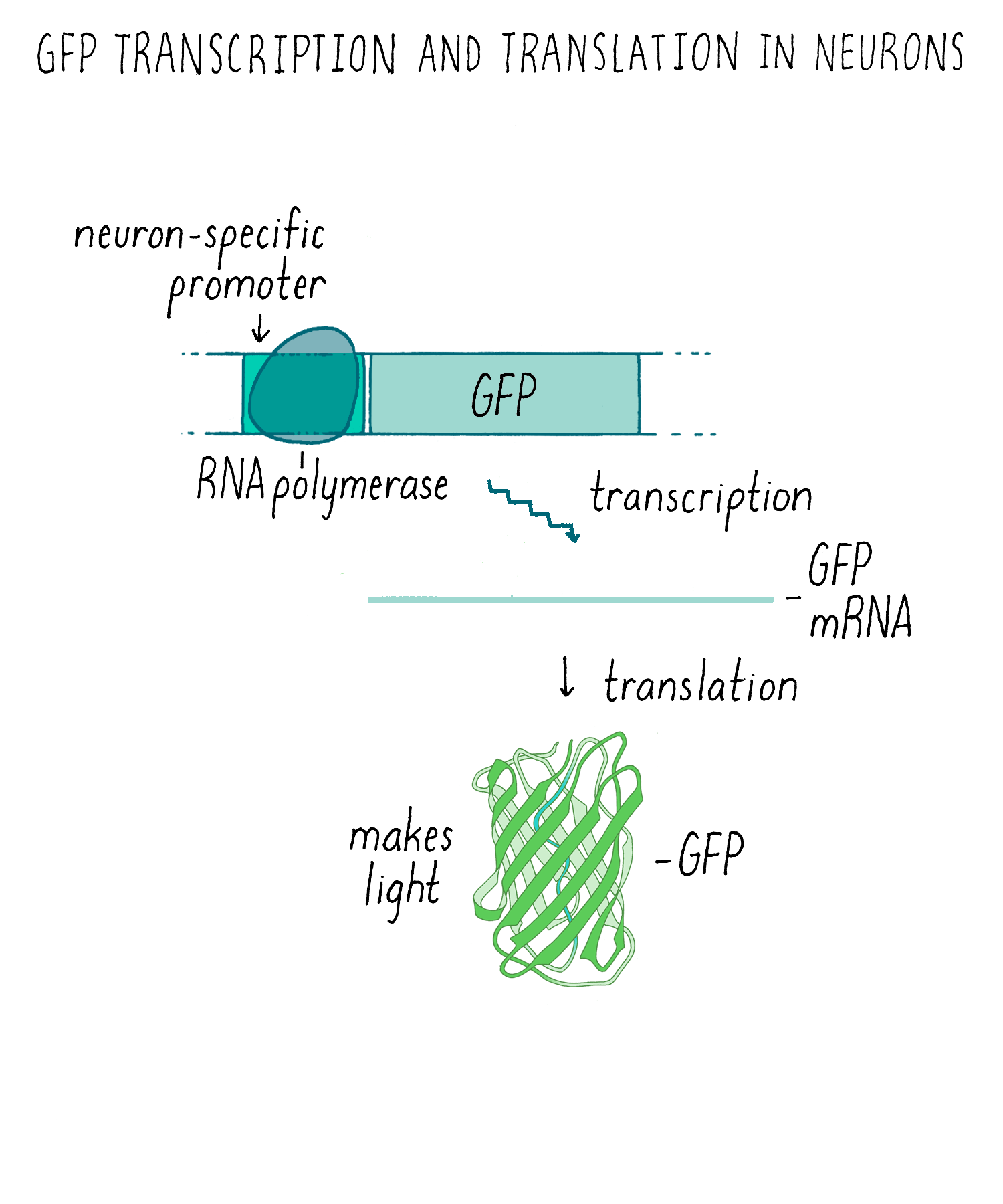
We then injected the GFP plasmid into the gonad of the worm where it becomes incorporated into gametes (both sperm and egg since C. elegans is a self-fertilizing hermaphrodite). After fertilization, the resulting animal can now express GFP (Figure 10). This procedure worked remarkably well. Finally, we could see specific neurons by GFP fluorescence and watch them in living animals.
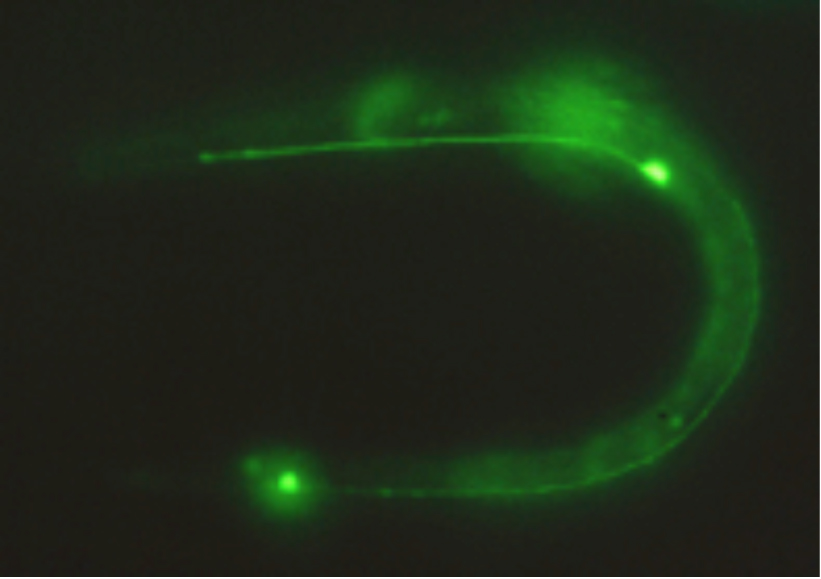
The actual fluorescence image of GFP expressed in touch-sensing neurons in a living C. elegans worm is shown in Figure 11.
The final paper also had additional comments on using GFP and some suggestions of possible future uses. Since we had already begun sharing GFP cDNA with other scientists (and their experiments had succeeded), we were also able to cite their unpublished work to indicate that GFP was going to be a useful tool in many organisms.
What happened next?
Experiments answer questions, but they also invite many more. We knew that we could express GFP to label cells in living animals (Figures 10 and 11). However, an additional exciting application was fusing the GFP DNA to DNA encoding another gene. Then, a “fusion protein” would be produced where the fluorescent GFP is added to the other protein (Figure 12). This fusion protein would allow one to visualize particular protein is localized in the cell, thus serving the same goal of immunofluorescence but without needing an antibody (which are time consuming to make). In addition, unlike immunofluorescence which kills the cells, a GFP-fusion protein could be imaged in living cells and one could make a movie to see if its localization changes over time. The first experiment of this type was done by my wife, Tulle Hazelrigg, who made the first GFP fusion protein and watched its movement in the developing fruit fly oocyte. Subsequently, hundreds of thousands of such GFP fusion proteins have been made and studied.
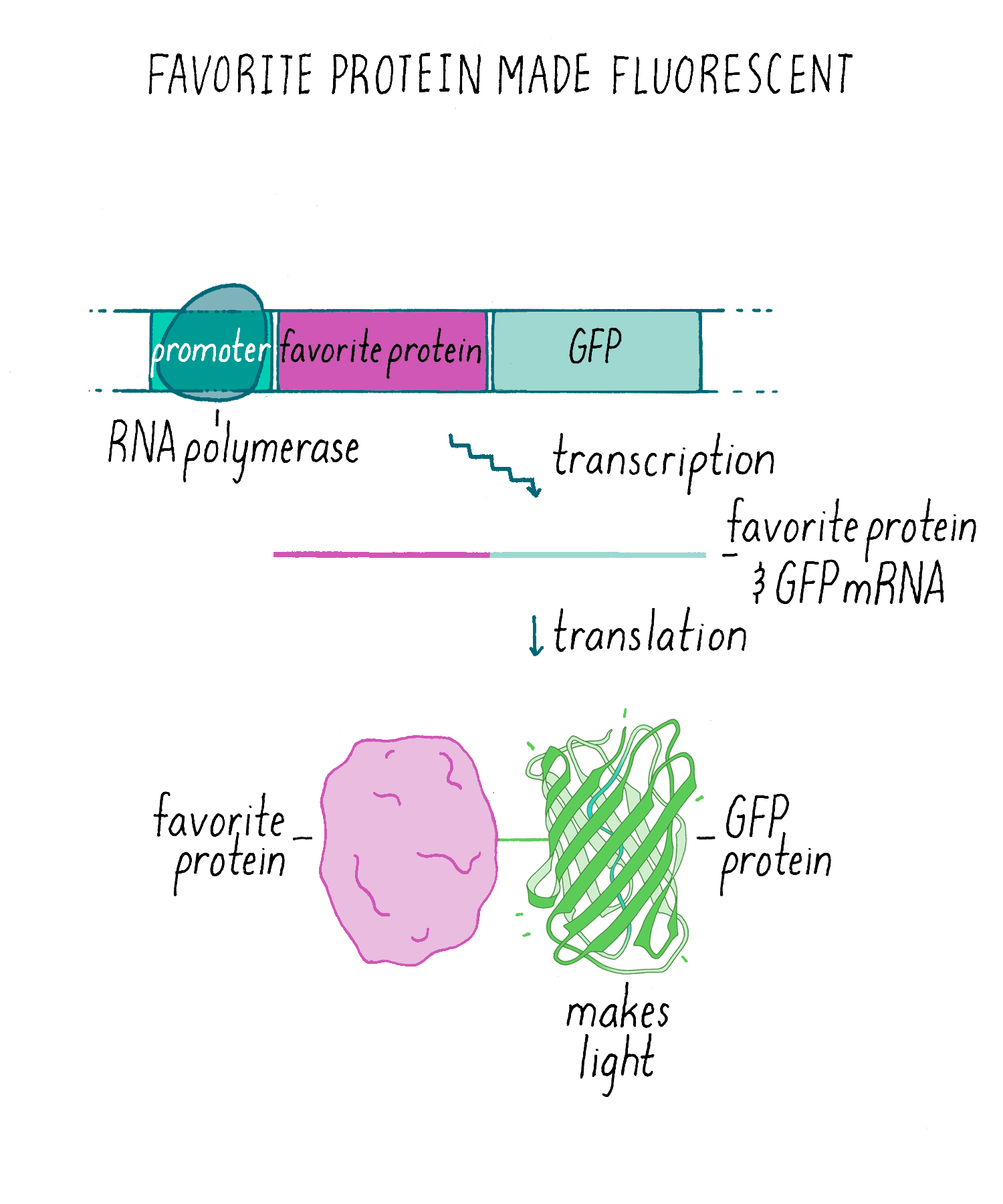
GFP fusion proteins were just the beginning. Like the mouse in Laura Joffe Numeroff’s book If You Give a Mouse a Cookie: if you give scientists one thing (GFP), they will always ask for more—more strongly fluorescing proteins and proteins that fluoresced in different colors. Indeed, soon after people started using GFP, scientists found new fluorescent proteins in coral and other organisms. Scientists also changed the DNA sequence of GFP and other fluorescent proteins so they fluoresced in other colors (e.g., yellow, blue, and cyan). Roger Tsien, who shared the 2008 Chemistry Nobel Prize with Shimomura and me, contributed greatly to the development of GFP as a tool for many uses in biology (see Tsien’s lecture on this topic on iBiology.org).
GFP and other fluorescent proteins have enabled an astonishing number of experiments. In 2014, 20 years after we had published our paper describing GFP as a biological marker, Chem Abstracts estimated that 160,000 papers used fluorescent proteins. As I first imagined, GFP has been used to label numerous organisms, cells, organelles, and proteins. This labeling has helped people look at gene expression and protein localization, follow biological events and disease processes, and search for mutants with abnormal cells or mislocalized proteins. I could not have imagined, however, the scale at which this work would be done. For example, GFP has been fused to all 6000 genes in yeast to look at the localization of every protein made in this organism. With GFP and its variants, scientists have now made countless movies of live cells (e.g., nuclear division in the fruit fly embryo; GFP fused to the protein tubulin that makes up the mitotic spindle, Video 3).
I imagined early on that GFP would be used in industry, and now pharmaceutical companies rely on fluorescent proteins to find new drug. And for entertainment, companies sell pet fish that glow in the dark, because of a transgenic fluorescent protein.
But GFP also has been used to do many things that I never imagined. Roger Tsien, for example, made a whole set of tools that can monitor cellular biochemistry, such as detecting changes in calcium levels, pH or protein phosphorylation, and other enzyme activities (see Tsien’s iBiology talk). Investigators in Japan coupled fluorescent proteins to silk fibroin to make fluorescent silk that they used to make clothing. Other investigators used GFP as part of a biological laser. In popular culture, GFP was evoked in the Ang Lee movie Hulk to explain why this superhero has green skin. One of the real joys of having introduced a new technology is being astonished by the remarkable new and imaginative uses that other people devise.
Closing Thoughts
I like to think about the GFP story as a counterexample to the usual story we hear about scientists and their experiments. In the standard narrative, scientists are geniuses who work alone and come up with a problem that has never been solved but by using the scientific method (develop a hypothesis, design an experiment to test the hypothesis, do the experiment, and demonstrate that the hypothesis is true), they can find an answer.
The GFP story, starting with the remarkable work of Osamu Shimomura, is nothing like this. Although his work started out with an interesting question (how are jellyfish bioluminescent?), throwing the preparation in the sink has never been part of the scientific method. (I have, however, heard of other scientists whose experiments worked after they used preparations that accidently spilled on the lab bench or even the lab floor.) And his most surprising discovery, GFP, came from the puzzling result that aequorin produced the “wrong” color light. I think the lesson here is that many if not most discoveries in science are accidental. In fact, the physicist Enrico Fermi once said:
“There are two possible outcomes [of an experiment]: if the result confirms the hypothesis, then you've made a measurement. If the result is contrary to the hypothesis, then you've made a discovery”
How does one make discoveries? Do lots of experiments and keep an eye out for the unexpected; they never come on cue. Sometimes you need some luck; I had no idea that I would work on GFP before that seminar I heard in 1989. Unfortunately, we sometimes don’t see the unexpected or don’t follow up an observation with new experiments. For example, in our GFP paper, I described a curious result I had obtained: GFP exposed to ultraviolet light would become “activated” and respond more strongly to blue light. I never followed up on this observation of “photoactivation,” but, fortunately, others did. Photoactivation proved to be a powerful method for tracking proteins in cells and also became the basis of a new microscopy method called “super-resolution microscopy” that led to a Nobel Prize in 2014.
I take several other lessons from our work with GFP.
First, science is often a collaborative effort. In fact, the reason that GFP has become such a profoundly useful tool is because so many people added their knowledge, time, and ideas to make it so.
Second, sometimes you have to ignore perceived knowledge (e.g., GFP cannot work on its own). Thus, ignorance, stubbornness, and curiosity are often very useful traits in the lab because they encourage you to test wild ideas. If you are too embarrassed to tell people about such craziness, you can always do the experiment over the weekend when no one else is around.
Third, sometimes sloppiness doesn’t hurt, as long as you realize what you are doing and are still rigorous in collecting and reporting your data.
Fourth, we should be studying all of life. We don’t know what new “GFPs” are out there, and we will never find them if we only look at a few favored biological organisms. “CRISPR-Cas9” (see Narrative on CRISPR by Barrangou and Key Experiment on CRISPR-Cas9 by Doudna) is another example of a wonderful gift from nature that, like GFP, generated a powerful new technology.
Fifth, basic or fundamental research is essential. GFP was discovered as part of investigation into a fascinating biological question that had nothing directly to do with human health or a patentable application. Nonetheless, GFP has been used to understand other biological problems, to investigate disease processes (not by looking in humans, but by looking in other animals), and has been used in the biotech industry. Basic or fundamental research is needed, because it allows us to learn more about our world and enables applications that we cannot even imagine now.
Dig Deeper
Dig Deeper 1: Cloning a Gene
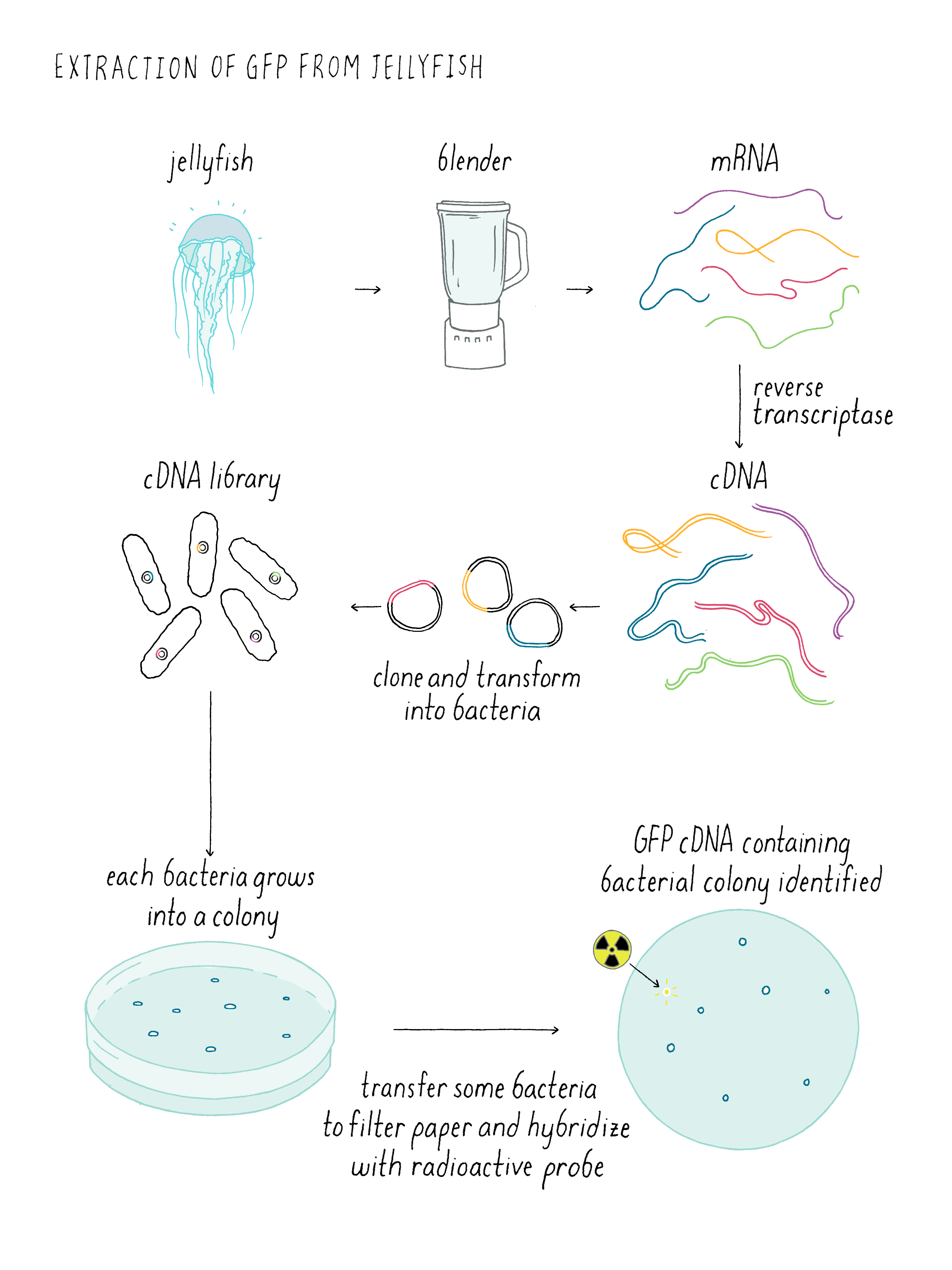
The cloning of the jellyfish GFP gene involved several steps (Figure DD 1). It was much harder to clone genes before our modern era when we have the sequences of many genomes. First, a complex mixture of mRNAs is isolated from the jellyfish. Normally, DNA is converted into mRNA by RNA polymerase. However, there is a trick for going backward and making DNA from mRNA using a special viral enzyme called reverse transcriptase (see White Board video on Reverse Transcriptase). With reverse transcriptase, DNA copies of all of the mRNAs (which are called complementary DNA (cDNA) can be made. These cDNA can be inserted into “vector” that can replicate inside of a bacteria and thereby amply the copies of the cDNA. Here shown are cDNAs inserted into a plasmid that could grow in E. coli. Doug Prasher used a virus vector called lambda, but the principle is the same. The collection of these plasmids (or lambda viruses) covering all of the jellyfish mRNAs is known as a cDNA library. When the library is added to bacteria under special conditions, the bacteria will incorporate the plasmids inside of them. Each bacteria tends to pick up just one plasmid; thus each bacteria clone makes a different cDNA.
Now one has to find the bacteria making a cDNA corresponding to the sequence of jellyfish GFP. The strategy is to make a radioactive DNA sequence that could bind to the complementary DNA sequence from the jellyfish DNA. First, filter paper is then placed on the petri dish with the bacterial colonies and then some of the bacteria are transferred to the filter paper; the positions of the bacterial colonies are preserved on the filter paper. The bacterial cDNA is heated so that the two strands “melt” and then when cooled down, one strand can form Watson-Crick base pairs with the radioactive cDNA probe, which is called “hybridization.” Since one can measure radioactivity, the hybridized radioactive DNA “points” to the location of correct bacteria carrying the sequence for the jellyfish DNA on the plate. By lining up the filter paper with respect to its original orientation on the plate, one can identify the bacterial colony that contains the cDNA of interest. Finally one scoops up these bacteria with a loop and grow the desired bacteria in liquid culture. Like finding a needle in a haystack, the bacteria carrying the cDNA of interest is identified and can be used to make lots of copies of the DNA. Hopefully, that cDNA contains the sequence of the entire gene. If not, one might have to find another bacteria with a bigger cDNA sequence. In fact, this is what Doug Prasher had to do to get the entire GFP gene from jellyfish.
References and Resources
Guided Paper
M. Chalfie, Y. Tu, G. Euskirchen, W.W. Ward, and D.C. Prasher. Green fluorescent protein as a marker for gene expression. Science 1994;263:802–805.
This is the paper that featured the Key Experiment described here.
This paper describes the discovery of green fluorescent protein (GFP), which allows scientists to monitor gene expression and protein localization in living cells. This research led to Chalfie receiving his Nobel prize in 2008.
DownloadReferences
- A. K. Corsi, B. Wightman, and M. Chalfie. A transparent window into biology: A primer on Caenorhabditis elegans. Genetics 2015;200:387–407
. An introduction to C. elegans biology.
- WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.177.1, Available at: http://www.wormbook.org.
Additional resources on C. elegans.
Resources
- Nobel Prize Web Site. Available at: http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2008/
A good place to start learning more about GFP is the Nobel Prize web site has popular, illustrated, and advanced descriptions.
- Osama Shinomura’s Nobel Prize acceptance speech. Available at: http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2008/shimomura-lecture.html)
A fascinating tale of his influential discoveries.
- Roger Tsien’s “Fluorescent Proteins” on iBiology. Available at: https://www.ibiology.org/talks/fluorescent-proteins/
A great overview by the late Nobel Prize winner Roger Tsien.
- Roger Tsien’s “Fluorescent Protein Indicators”. On iBiology. Available at: https://www.ibiology.org/talks/fluorescent-protein-indicators/
An in-depth presentation on biochemical reporters using fluorescent proteins by the late Nobel Prize winner Roger Tsien.
- Green Fluorescent Protein Video. Available at: https://www.youtube.com/watch?v=wxf4a4SX84A
This video briefly describes how GFP was discovered as well as some of the fascinating uses and discoveries that have been made using GFP technology.
- Excitation and Emission of GFP. Available at: https://www.biotek.com/resources/technical-notes/excitation-and-emission-of-green-fluorescent-proteins
This website describes the excitation and emission profiles of GFP using graphical representations as well as excitation and emission tables and charts.
- Visualizing Gene Expression Patterns Slide Show. Available at: https://www.hhmi.org/biointeractive/visualizing-gene-expression-patterns
This slideshow outlines different ways scientists can visualize mRNA and proteins expressed throughout an organism.