The Laws of Inheritance
A Journey from Mendel’s Abbey to Iceland
Shirley Tilghman
 Shirley Tilghman
Shirley Tilghman
Shirley M. Tilghman has been a professor and developmental geneticist in the Department of Molecular Biology at Princeton University since 1986. She also served as the 19th president of the University from 2001 to 2013. In 1996, she received the President’s Award for Distinguished Teaching, which recognized her many roles in teaching and mentoring. She is currently teaching an undergraduate seminar that examines the intersection between modern genetics and public policy.
Summary
Modern biology rests on two monumental 19th century discoveries – the laws of inheritance by Johann Gregor Mendel and natural selection by Charles Darwin. Mendel was that rarest of modern scientists – a solitary genius. Toiling alone in his garden studying various traits of garden peas, Mendel applied exquisite experimental design with rigorous quantitation to show that traits are inherited from parents as discrete units, which we now call genes. Traits come in pairs – one from each parent. Traits can be dominant, in which case their effect is always evident, or recessive, in which case they disappear when paired with a dominant trait. Mendel found that certain traits are not inherited together, but segregate into the next generation independently of one another. Mendel’s laws proved to be universal for all plants and animals. In humans, traits like freckles or diseases like cystic fibrosis obey Mendel’s rules because they are governed by single genes. However, most human traits arise from actions of many genes that can be influenced by the environment. Understanding such complex traits requires studying large numbers of individuals. Iceland provides a powerful modern genetic “laboratory” that allows scientists to identify the multiple genes underlying complex human traits and may have profound consequences for treating diseases such as cancer and behaviors such as addiction.
Learning Overview
Big Concepts
A living organism’s traits are controlled by its genes, and the most fundamental rules for how an offspring inherits genes from its parents and how those genes give rise to traits were established by Mendel. Mendel’s experiments uncovered the concepts of “dominance” and “recessive” and the independent segregation of genes. We now know that some traits and diseases are controlled by single genes (monogenic), whereas others are governed by several or even hundreds of genes (multigenic).
Bio-Dictionary Terms Used
Amino acid, chromosome, cross, enzyme, gene, fertilization, stop codon
Terms and Concepts Explained
Allele, co-dominance, diploid/haploid, dominant/recessive, F1 and F2 generation, genetic cross, genome-wide association study (GWAS), Mendelian inheritance, multigenic trait, ovule, phenotypes, pollen, sickle cell anemia, single nucleotide polymorphism (SNP), stamen, traits
Introduction
-
What set Mendel apart from his contemporaries was his systematic and quantitative approach to studying biology; these characteristics proved crucial for in his quest to uncover the basic rules for inheritance.
Part I: Journey to Discovery - Mendel’s Peas and the Laws of Inheritance
-
Great science begins with an important question. Mendel posed a truly fundamental one – how is the information that determines specific traits passed from one generation to the next?
-
Mendel’s experiments succeeded because of a powerful combination of extraordinary insight, immense persistence, and a little luck. He chose to study garden peas and their simple well-defined traits like petal color and seed shape. Those choices proved to be critical to his success. When he tried to reproduce the findings with another plant, the results proved too complex to interpret.
-
Mendel began by studying the inheritance of seven traits one by one. In this way, he reduced the number of factors that were likely to vary in each experiment, which made it easier to see coherent patterns in the data.
-
Mendel anticipated things that could go wrong and potential sources of error or confusion in his experiments, and he controlled for them. For example, he realized that random pollination from the environment would mean that he could not be certain of the identity of the parents of his cross. Likewise before conducting his crosses, he confirmed by breeding over multiple seasons that each parental plant “bred true” and produced highly reproducible offspring.
-
Mendel was a mathematician and appreciated the importance of quantitation. He also appreciated that experiments must be repeated many times for a clear pattern to emerge from the data. He realized that small numbers could potentially mislead him, as one might encounter if one wanted to determine the frequency of heads or tails with a just a few flips of a coin.
Part II: Knowledge Overview - The Fundamentals of Inheritance
-
Mendel discovered that simple traits are inherited from parents as single discrete units which we now refer to as genes.
-
The sequence of a gene is not identical from individual to individual. The different variants are called “alleles” that can result in differences in the way in which the trait is displayed – the “phenotype.” Alleles are dominant, recessive, or co-dominant. When a dominant and recessive trait is inherited together, the phenotype associated with the dominant trait will prevail. If two co-dominant alleles are co-inherited, the resulting phenotype is somewhere between the two parental phenotypes.
-
Simple traits are inherited independently of one another (unless they are encoded near one another (linked) on the same chromosome).
-
Many traits in species, including humans, are not monogenic, that is, they are not governed by the action of a single gene, but by the combined action of multiple genes (multigenic).
Part III: Frontiers - Uncovering Genes Involved in Multi-Genetic Traits
-
Identifying the genes that govern complex traits cannot be studied using Mendel’s approach. Scientists have developed an approach called Genome-Wide Association Studies, which identifies correlations between gene variants and traits.
-
GWAS exploit natural variation in the sequences of DNA in human populations – called single nucleotide polymorphisms (or SNPs) – and use them to act as landmarks of specific regions in the genome.
-
Large populations of affected and unaffected individuals are sampled for thousands of SNPs, and statistical correlations are sought between individual SNPs and phenotypes.
-
The position of the SNP in the genome directs the scientists to the genes in the immediate locale that might be responsible for the phenotype.
-
We will explore a GWAS investigation performed in Iceland, which identified genes that contribute to eye and skin color as well as other traits.
-
GWAS are currently being used to study complex human diseases that have genetic as well as environmental contributions, including cancer, addictive behavior, obesity, and heart disease.
Closing Thoughts
-
Human traits involve complex networks of many genes. The actions of genes are also influenced by the environment; thus, genes are not destiny. Understanding gene networks and environmental influences constitutes one of the great challenges of the 21st century.
Guided Paper
Mendel G. 1866. Experiments in Plant Hybridization. Translated by Scott Abbott and Daniel J. Fairbanks. 2016. Genetics 204: 407–422. https://doi.org/10.1534/genetics.116.195198
This is a modern translation of the classic paper by Mendel. In addition to defining for the first time the concepts of dominant and recessive traits, Mendel’s research uncovered the laws of inheritance by which traits are passed from one generation to another.
DownloadActivity
Explore the beautiful shapes of pollen yourself with the Foldscope Activity on Pollen.
Introduction
Anyone who has sat around a family gathering, noticing resemblances between parents and children, uncles and cousins, and grandparents and grandchildren, has engaged in genetic analysis. You may have noticed that your cousin John is tall, just like his father and mother, or that almost everyone in that family has brown eyes. Yet John’s brother Alex may be quite a bit shorter and have bright blue eyes. You may notice that when the family members cross their arms, they all put the left arm over the right one, even Alex the outlier. What you are doing is asking how “traits” – the characteristics that describe our appearance, behavior, disease susceptibility, and physiology – are passed from one generation to the next. Are there rules or is it simply a random chance?
The individual who provided the first definitive answer to this question was Johann Gregor Mendel (1822-1884), an Augustine monk living in a monastery in Brno, Austria (now the Czech Republic). Everyone who has taken an introductory biology course has probably encountered the discoveries of Mendel and his peas, presented as a series of straightforward experiments with clear-cut results. Calling myself a geneticist for almost 40 years, I certainly thought I knew what Mendel had done! But a few years ago, as I was preparing a course on the greatest experiments in biology, I read Mendel’s original paper for the first time (and you can too - see Mendel’s Guided Paper). I was gobsmacked! In that paper, Mendel displays every characteristic of a great scientist: laying out his motivation and the preliminary groundwork; choosing his organism; taking painstaking steps to eliminate sources of error; making careful observations and converting them into data; detecting patterns in the data; interpreting the patterns to reach general conclusions; and acknowledging failures.
Mendel’s paper reads like a “How to” manual for conducting great science, still relevant for modern times. In this Narrative, I will try to convey the wonder of scientific discovery by describing Mendel’s beautiful experiments and what made them great in The Journey to Discovery.
Like Charles Darwin, who together with Mendel is the founding father of modern biology, Mendel was a 19th century naturalist – someone who observed the natural world and tried to draw conclusions about how it works. However, Mendel did not set out to be a biologist. He was born into a farming family in Austria and would have concluded his education after primary school except for a teacher who recognized promise in the young man and recommended that he continues his education. In college, he studied mathematics and physics, and after his graduation, he joined the Augustine religious order. This decision was not unusual at that time for a scholarly young man. The Augustinians were intellectual leaders of their day, and the St. Thomas Abbey housed extensive library and experimental facilities.
After completing his religious studies, Mendel took and failed an exam to obtain a teaching certificate because he lacked sufficient knowledge of botany! The monastery sent him to the University of Vienna to continue his studies, and it was there that he encountered his mentor Franz Unger, who introduced him to the study of species. When he returned to the abbey in Brno to begin his own experiments into the nature of hybrids, he commented that “It requires indeed some courage to undertake a labor of such far-reaching extent; this appears, however, to be the only right way by which we can finally reach the solution of a question the importance of which cannot be overestimated in connection with the history of the evolution of organic forms.”
What set Mendel apart from his predecessors was the systematic and quantitative way he went about tackling the principles of inheritance. He approached the problem as a mathematician, understanding that small numbers can fluctuate and that if he was to uncover laws governing inheritance, he would need to repeat his experiments many times. This conviction proved to be critical to his success in uncovering the laws of inheritance.
It is the dream of every scientist to uncover laws and principles that are generalizable and apply, not just to one organism, but to all living things. Mendel’s discoveries certainly fall into this category, but sadly, his achievements were not recognized in his own lifetime. It was not until well after his death, when his experiments were “rediscovered” by three European scientists at the turn of the 20th century, that it became clear that his laws were universal. Indeed, as we will see in the Knowledge Overview of this Narrative, Mendel’s laws of inheritance apply to humans as well, and we will revisit Mendel’s laws with our modern understanding of DNA and chromosomes.
We will also see that Mendel was extremely lucky in his choice of traits to study in his peas, for when it comes to human traits, it is more often the case that traits and phenotypes are governed by many genes, making the study of inheritance patterns much less straightforward than Mendel observed. That is why the observations you make around the dinner table are often more confusing than clear. We will see in the Frontiers section how scientists today use Genome Wide Association Studies to hunt down the genes that underlie the charm of freckles or the unfortunate consequence of schizophrenia. This is where the exciting future of modern genetics lies.
Part I: The Journey to Discovery —
Mendel’s Peas and the Laws of Inheritance
The Problem
Mendel was not the first person to question how the properties of living organisms are passed from one generation to the next. At the time of his studies, European “naturalists,” as scientists were then called, were absorbed in debates surrounding the appropriate classification of organisms, whose number and characteristics were thought to be fixed according to the Judeo-Christian Book of Genesis. It was generally believed that traits of offspring were often a blend of the traits in the parents, but that over many generations a hybrid would always revert to its original form. A few courageous individuals such as Mendel’s professor in Vienna, Franz Unger, opposed that view and proposed that entirely new species can arise over many generations through gradual change and hybridization between existing species – what Charles Darwin was to demonstrate definitively a few years later.
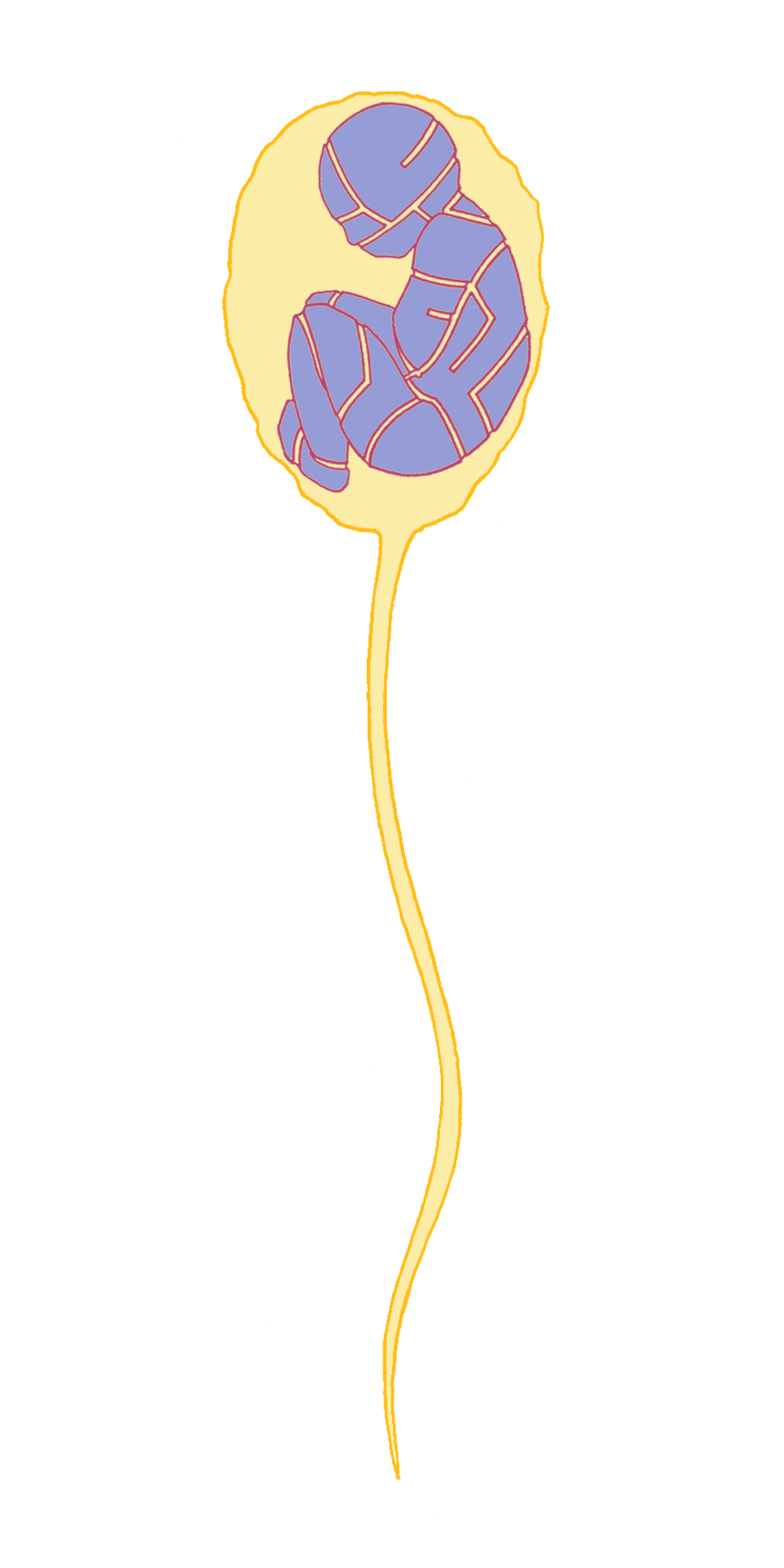
Another orthodoxy of the time that Mendel challenged was championed by Edward Fenzl, the Director of the Vienna Botanical Gardens. He believed that plant embryos were already fully formed (although microscopic) in pollen grains and that the ovule (the seed) only provided a rich environment in which the embryo could grow. This “spermist” theory was also applied to animals and humans, postulating that there was a pre-formed person contained within the sperm (Figure 1). This was consistent with the prevailing view that there was divine order in nature and that all living things were fixed for all time.
Mendel set out to determine whether there were rules – or laws – that governed the behavior of how "characters” (Mendel’s term for what biologists today call "traits” or "phenotypes”, referring to a particular physical or chemical feature of an organism) are passed down from one generation to the next. Today it is hard to imagine how ambitious this was, as Mendel began with no knowledge that traits are encoded by genes that are assembled in parallel into long polymeric molecules of DNA called chromosomes. He had no clues to follow, as there was virtually no pre-existing scientific literature on this topic. Nor was he aware when he began his experiments of the thinking of his contemporary Charles Darwin, who was just beginning to work out his theory on the origin of species. He did not have the advantage of working in a large university or a research institute full of fellow scientists to challenge and improve upon his ideas. Instead, he worked alone in a small garden beside the monastery wall.
Mendel was that rare breed – the lone genius.
Clues
Clue 1: The choice of peas
The first critical decision that any biologist faces is the choice of an organism to study. Mendel’s original idea was to study mice, and he began breeding them in his monastery bedroom, but the Abbot vetoed the idea because he was opposed to a monk studying sex! Clearly, the Abbot was not a biologist, for he didn’t appreciate that what Mendel was to investigate next was the sex life of peas!
For his work, Mendel chose the garden pea, Pisum sativum. Mendel recognized a number of advantages of starting with peas. Over centuries gardeners had bred them to display many varying properties such as height, seed shape, seed color, pod shape, pod color, and the position of flowers on the stems. However, he did not trust the gardeners from whom he received the varieties. He spent two long years cultivating each variety before beginning to hybridize them (for more information on plant breed, see the Narrative on Plant Genetics by Ronald). He was ensuring that the plants "bred true” – that each plant within a variety was identical to every other. This guaranteed that any differences he observed in subsequent hybrids were due to the hybrid cross, and not to some inherent variability in the original variety. This is an example of a scientist anticipating a potential source of error and eliminating it.
Explorer’s Question: Mendel decided to study a number of traits that varied in peas. Why?
Answer: Mendel ultimately examined the inheritance of seven different traits that were easily measurable and sufficiently different from one variety to another. He wanted to follow the inheritance of more than a single trait to ensure that his conclusions from studying one trait were applicable to others. He was looking for universal laws.
Explorer’s Question: Peas are easy to grow in large numbers, with multiple crops per year. Why was this important to Mendel?
Answer: Mendel anticipated that in genetics, numbers would matter. If you are going to study traits as they pass from one generation to the next, the more organisms you can grow, and the more generations you can study in a year can make a big difference. Think about the challenge of studying genetics using elephants!
Explorer’s Question: Mendel knew from the work of others that hybrids (the offspring of two phenotypically distinct parents) between pea varieties were fertile. Why was this important?
Answer: Mendel anticipated that he was going to study multiple generations of peas, so the hybrids needed to be fertile. Many types of hybrids, for example, the hybrid mule that arises from a mating between a horse and a donkey, are sterile.
Explorer’s Question: Peas are usually self-fertilizing and the shape of its flower inhibits cross-fertilization. Why does this matter?
Answer: Mendel wanted to have complete control of experiments, which meant being in control of plant breeding. He grew his plants in both a greenhouse and in a nearby field. If his plants could be randomly fertilized by pollen carried by insects or blown in the wind, he would have had no way to verify the identity of the parents of his hybrids, and his results would not have exhibited clear patterns of inheritance. By choosing a plant that made unintentional cross-fertilization unlikely, he anticipated a potential confounding variable and greatly reduced it. Controlled hybridization is also discussed in the Narrative on Plant Genetics by Ronald.
As you can see from above, Mendel’s choice of the pea as an experimental organism was well thought through. However, as you will see later that, it was also lucky. If he had started with the bean plant, it would have been much more challenging for him to come to derive his simple laws. As the cliché goes, sometimes it is better to be lucky than good. Mendel, of course, was both.
Mendel made one more critical decision in setting up his experiments. Unlike his predecessors, who chose to study plants that differed in multiple traits (which made the task of identifying patterns of inheritance difficult), Mendel chose to systematically study the inheritance of each of seven traits one at a time.
The Discovery: Rules of Inheritance
Mendel’s First Discovery – Traits are Inherited as Discrete Pieces of Information
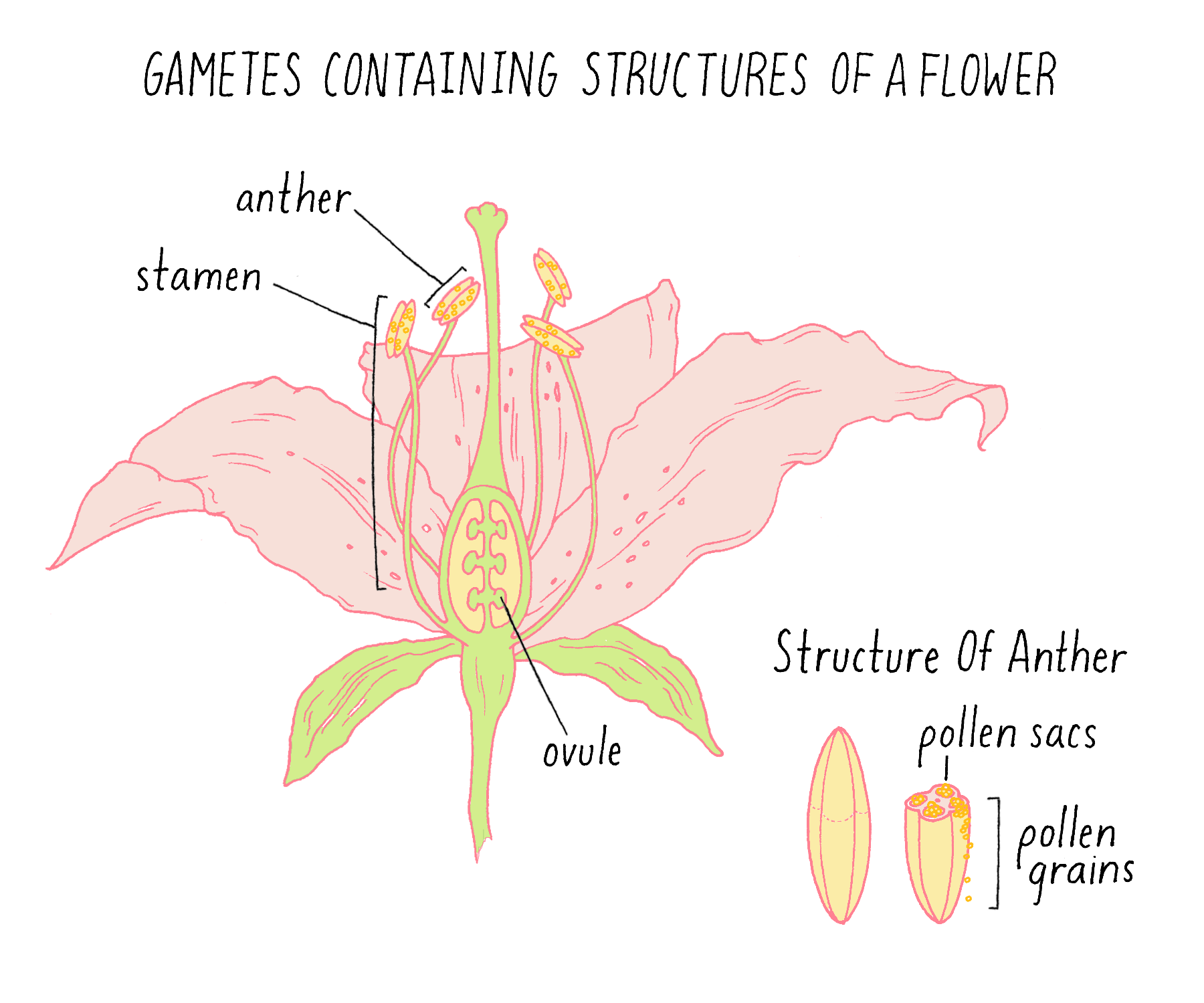
Once he knew that each variety of his peas bred true, Mendel began to set up crosses between varieties, in which pollen from one variety was used to fertilize the ovule of a variety that differed in one of the seven traits. Pollen and ovules are the plant terms for gametes – the equivalent of sperm and eggs in animals (Figure 2). (If you want to explore the beautiful shapes of pollen yourself, see the Foldscope Activity and Exercise on Pollen.)
Because plants are self-fertilizing and can produce both pollen and ovules, Mendel had to meticulously remove the stamens from immature plants of one variety so that they could not produce their own pollen. He then fertilized those de-pollinated plants with pollen from a variety that differed in one easily measurable trait. Tall plants were fertilized with pollen from short plants; plants with round seeds were fertilized with pollen from plants with wrinkled seeds. In all, he set up seven different crosses (a cross refers to a deliberate breeding between two individual plants or animals). The mature seeds that developed from the artificially fertilized plant were then planted, and the traits of the progeny plants (referred to as F1 progeny) that Mendel observed are shown in Figure 3.
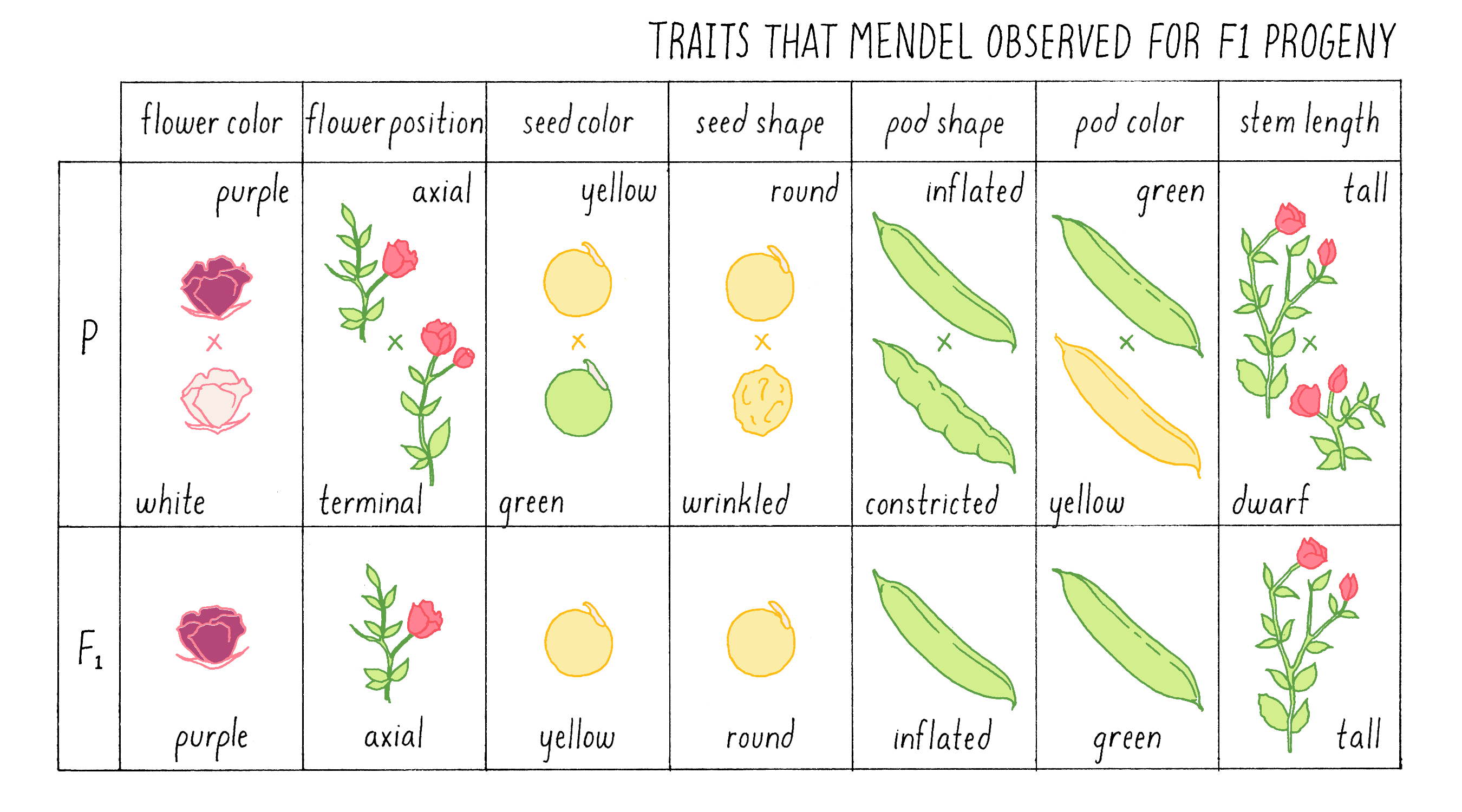
Explorer’s Question: What strikes you about these results? What would you have expected?
- Only one of the traits in the parental varieties appeared in the F1 progeny
- There were no intermediate phenotypes observed
- There was no consistency in the results
- The results could not have been predicted in advance
- All of the above
Answer: If you chose 1, 2, and 4, you were correct. The results provided Mendel with his first clue to the rules of inheritance. In each cross, only one of the two parental traits was expressed, and there were no intermediate or novel phenotypes. In the tall x short cross, for example, all the F1 plants were as tall as the tallest of the parents; indeed, some were even taller.
Mendel’s observation that there were no intermediate traits was completely unexpected. The current thinking of the time was that traits exhibited by hybrids were blends of the traits in the parents. For example, if one crosses poodles and golden retrievers, the offspring superficially appears to be a blend – the adorable goldendoodles. Overturning dogma (no pun intended) is never easy, but Mendel had numbers on his side.
Mendel referred to the character that was displayed in the F1 plants as dominant and the trait that "disappeared” as recessive, terms that we still use today to describe the behavior of genes in genetic crosses.
Mendel was careful to show that his findings were consistent and reproducible; in total, he performed 287 independent fertilizations of his varieties. By this point, he had outgrown the greenhouse and the small garden adjacent to the abbey wall that the Abbot had assigned to him, and he was consuming more and more land around the abbey.
For the next (F2) generation, Mendel fertilized one F1 plant with pollen from an identical F1 plant, a process called intercrossing or self-fertilization (see also the Narrative on Plant Genetics by Ronald). When the seeds that resulted from this cross, which are called F2 progeny, were grown to maturity, he observed and quantified their phenotypes for almost 20,000 plants in total! see Figure 4.
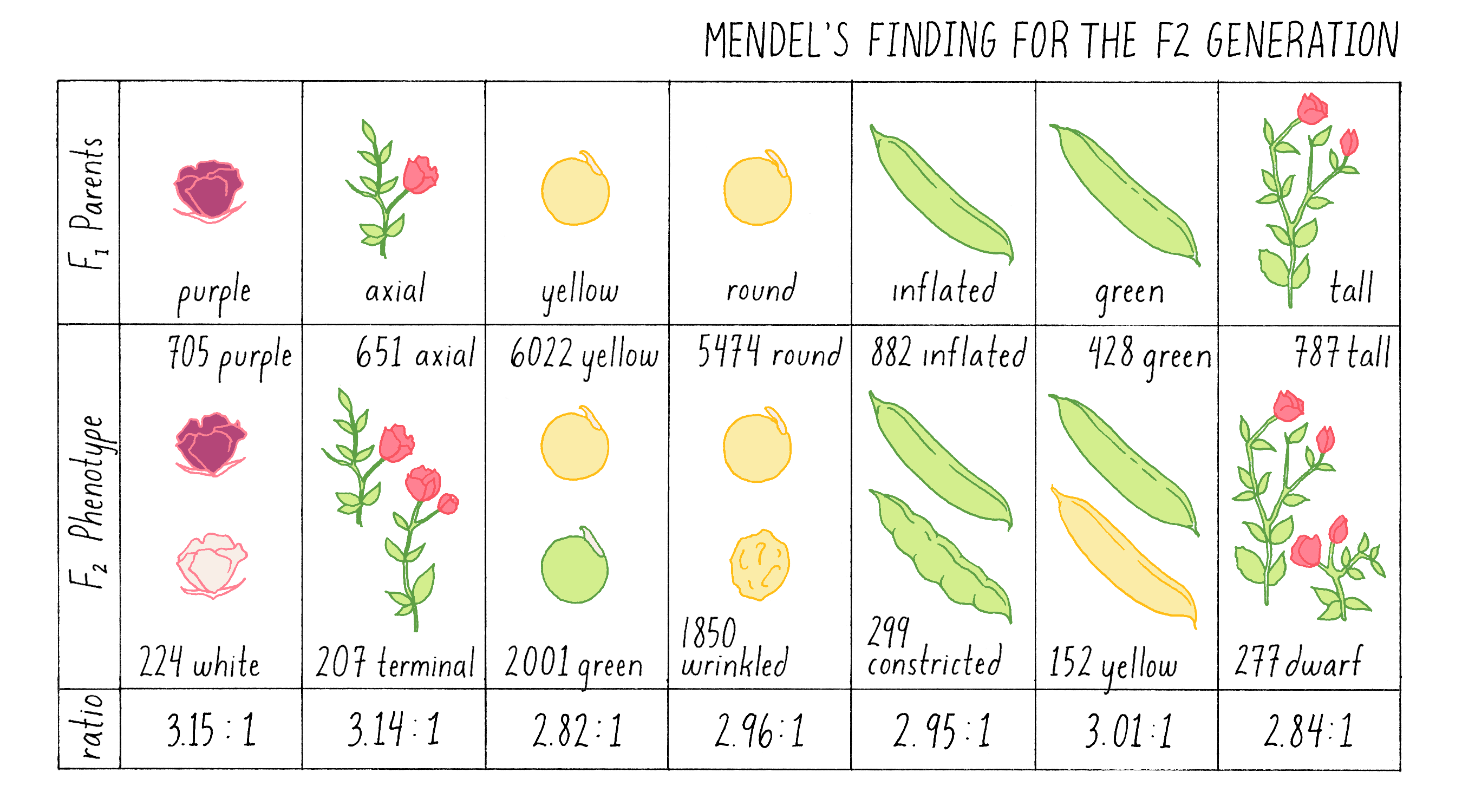
Explorer’s Question: What is most striking about these findings?
- The number of plants that Mendel analyzed
- The reappearance of the recessive trait in each case (e.g., short)
- The similarity in the ratios of dominant to recessive traits (~3:1)
- No ratio was exactly 3:1
- All of the above
Answer: If you answered all of the above, you would be correct. The reappearance of the recessive trait consistently occurred in each intercross, unchanged in character despite its disappearance in the F1 parents. Furthermore, it reappeared in a predictable manner – on average in one quarter of the plants.
Mendel’s initial concern about the fluctuation of small numbers was fully warranted. For example, if he had only intercrossed the tall F1 plants, and obtained the ratio of 2.84:1 of tall to short plants, he would not have been justified in rounding up to the critical 3:1 ratio. It was only when he had examined almost 20,000 plants in seven separate intercrosses that he could see that the ratios consistently approached an average of 3:1, just as a heads-versus-tails coin toss will only approach the precise average of 50:50 with many rounds of tosses. Anyone who has called a coin toss knows that at each toss, there is a 50:50 chance that the coin will land heads-up. Even after 5 consecutive tosses in which heads are called, the 6th toss has the same 50:50 chance of being heads. If you judged the probability of heads versus tails after only 5 tosses, you would be deceived into thinking that there was a 100% probability of heads. Only if you continued to toss the coin over and over again would the ratio of heads to tails approach the true 50:50 average. The behavior of traits in Mendel’s crosses behaved exactly the same – small numbers can be deceiving, and none of his crosses produced the exact 3:1 ratio. It took an extraordinary effort to convince himself – and ultimately the scientific community – that the 3:1 ratio was authentic and telling him something crucially important about the inheritance of traits.
Collectively, these results told Mendel that traits occur as discrete pieces of information (which we now call genes) that appear (dominant) or disappear (recessive) in F1 hybrids. Mendel proposed that a trait like "tall” was governed by what he called a dominant "character T” (recall that Mendel did not know about genes or DNA) and short was governed by a recessive "character t” (Figure 5; note: the upper case character for dominant trait and lower case for recessive is still used today). When a plant has only T traits, it is tall, but when it combines both characters in F1 hybrids (T and t), it is tall due to the dominant nature of T. But when two “Tt” plants are intercrossed, on average, one quarter of the progeny are “tt” in character, generating plants with the short phenotype.
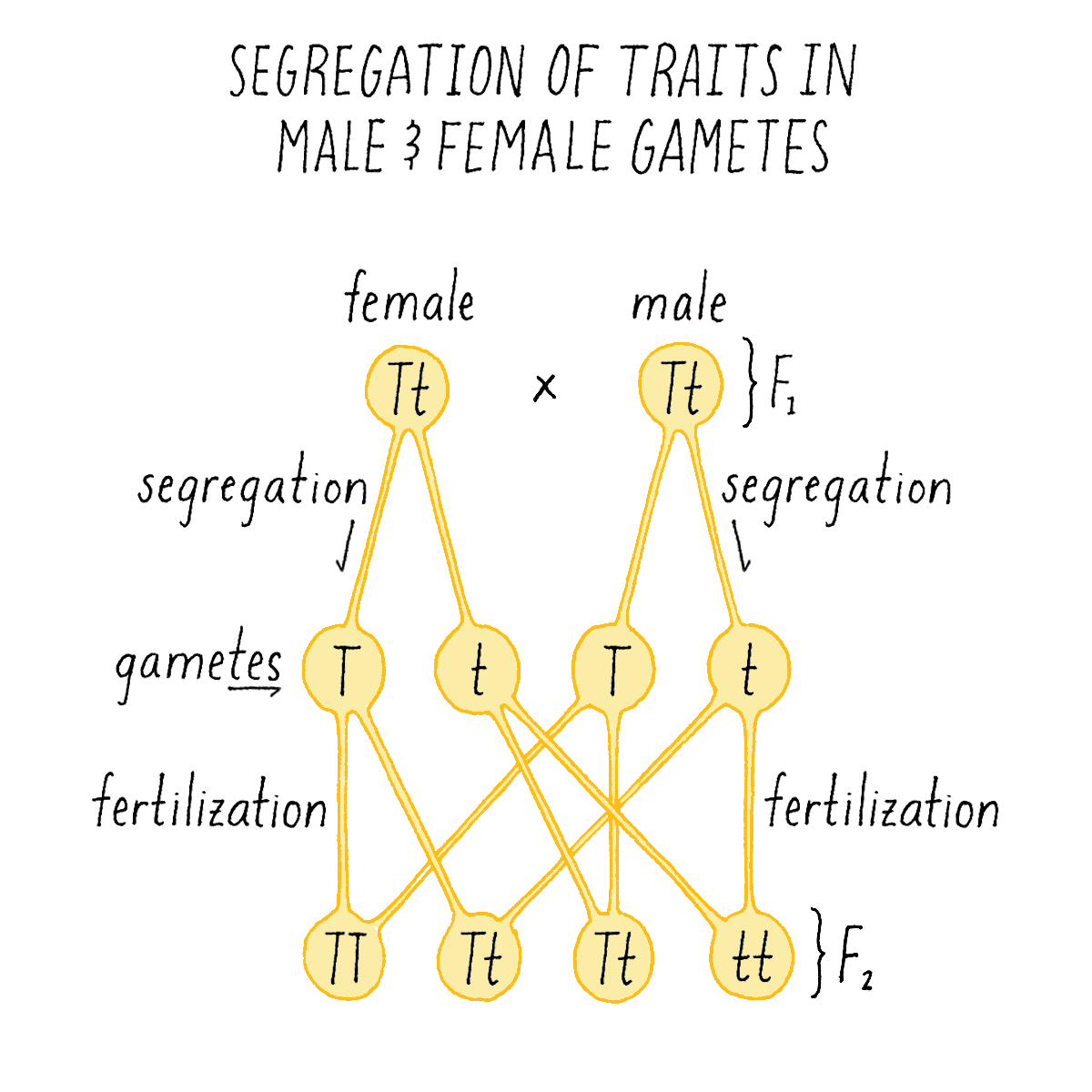
Mendel proposed that the 3:1 ratio of F2 traits could be explained, if each F1 parent contained two characters (T and t) that had been inherited from the parental varieties, and that the gametes (the ovule or pollen) they produced contained one character or the other, but not both. When the gametes combine to produce the F2 plant, they do so randomly so that, the offspring will appear in a ratio of 3 three-tall plants (TT, Tt, tT) for every short plant (tt) (Figure 5).
In Mendel’s own words:
"Since the members of the first generation spring directly from the seeds of the hybrids, it is now clear that the hybrids form seeds having one or other of the two differentiating characters, and of these one-half develop again the hybrid form, while the other half yield plants which remain constant and receive the dominant or the recessive characters in equal numbers.”
From this observation, Mendel disproved the spermist theory that all information for future offspring was contained within the sperm head. The ovule contributes equal amounts of information as the pollen – not just a fertile environment for growth.
Like every good scientist, Mendel was not content to simply interpret his experiments and propose theories to explain his findings. Theories are only useful if they can be tested. Mendel tested his own theory by choosing at random a large number of F2 plants and self-fertilizing them.
Explorer’s Question: By considering the four different F2 genotypes in Figure 5 (TT, Tt, tT, tt), what phenotypes would you predict Mendel observed in the progeny of the self-fertilized F2 plants?
- 1/4 would produce only tall plants, 1/4 would produce only short plants, and 2/4 would produce tall and short plants in a ratio of 3:1
- All the plants would be tall
- All the plants would be short
- The plants would be 50:50 tall and short.
Answer: The correct answer is 1. Mendel performed this test and confirmed that 1/4 produced only tall plants (TT), 1/4 produce only short plants (tt), and 2/4 (Tt) produced tall and short plants in a ratio of 3:1.
Mendel’s Second Discovery – Independent Segregation of Traits
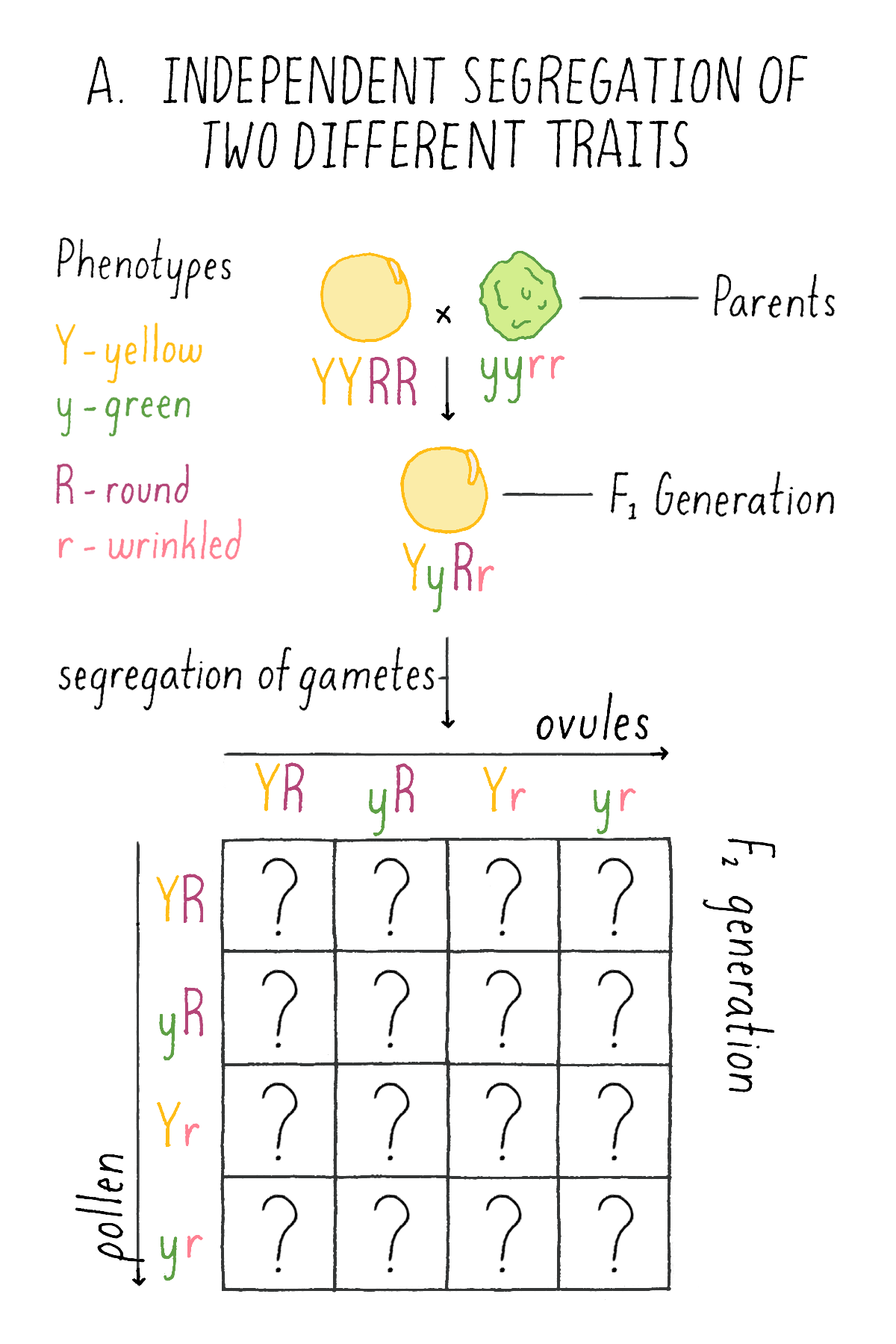
In the first set of experiments, Mendel systematically examined the patterns of inheritance of one character at a time. He next asked how two different traits would behave in hybrids. To explore this question, he crossed plants with round (R) and yellow (Y) seeds (two dominant traits) with another variety that had wrinkled (r) and green (y) seeds (two recessive traits; labeled with the same letter, but lower case, as the dominant trait). He wanted to know whether the traits of shape and color behave as a “linked” or “unlinked” pair – i.e., would they always appear together in future progeny, or would shape and color appear separately in the next generation?
Explorer’s Question: Based on the earlier experiments, what would you predict the phenotype of F1 hybrids in a cross between YR (Yellow, Round) and yr (green, wrinkled) plants to be?
- green and round
- yellow and round
- green and wrinkled
- yellow and wrinkled
- None of the above
Answer: The correct answer is 2 because Round and Yellow behave as dominant traits in the cross (see Figure 3).
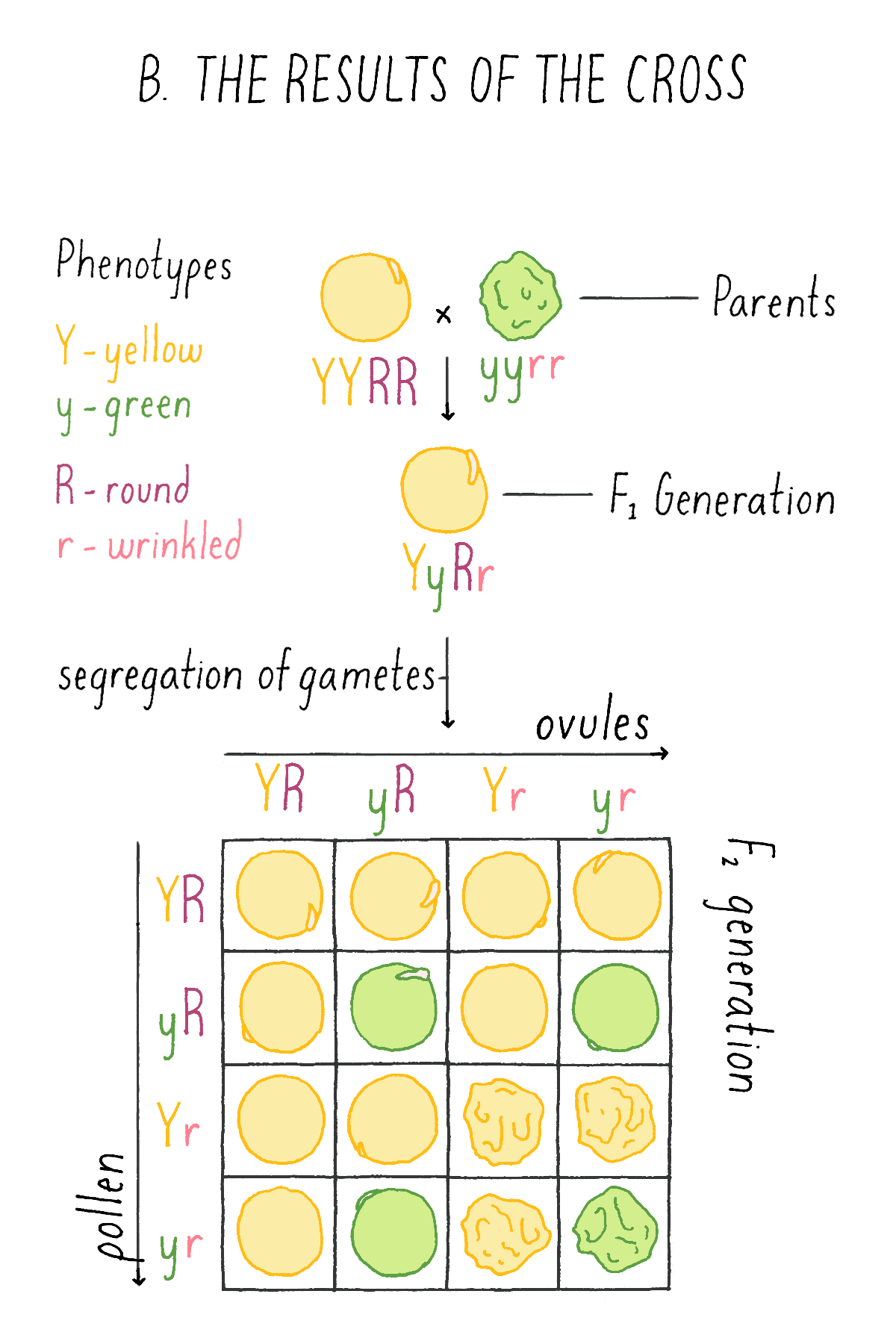
When F1 hybrids were intercrossed (self-fertilized), Mendel observed the following characteristics in their progeny:
- 315 yellow-round
- 108 green-round
- 101 yellow-wrinkled
- 32 green-wrinkled
Explorer’s Question: What do you notice about some of the progeny that makes them different from both their parents and grandparents?
Answer: The parents (F1) and one of the grandparents were both Yellow and Round (YR) and the other grandparent was green and wrinkled (yr). But now the F2 shows two new phenotypes – green-Round and Yellow-wrinkled.
This experiment by Mendel revealed that the two traits are not inherited together but behave independently of one another. Another striking observation was the finding that the four categories of phenotypes did not appear in equal proportions, but in a ratio of approximately 9:3:3:1. Mendel repeated many intercrosses using new combinations of the original varieties and consistently came up with the same 9:3:3:1 ratio!
What did this clue reveal to Mendel about the mechanism of inheritance? Mendel concluded that each of the gametes of the plants – the ovule and the pollen – harbored the discrete information for each character – i.e., either the round (R) or wrinkled (r) seed shape and either the yellow (Y) or green (y) endoderm. And, given the independent behavior of the shape and color phenotypes, each ovule and pollen will randomly contain one of the two characters for seed shape and one of the two characters for endoderm color. Thus, YR is as likely as yR, Yr, or yr.
Explorer’s Question: See Mendel’s experiment in Figure 6A, and see if you can assign genotypes and phenotypes to each class of progeny that would explain the 9:3:3:1 ratio. For example, in the top left box, a combination of a YR ovule with a YR pollen is a YYRR genotype and the corresponding phenotype is yellow and round.
What happened next?
Mendel was not content to discover laws that applied to peas alone, but like the best scientists before and after him, he was interested in discovering laws that were generally applicable to all living organisms. However, when he repeated his experiments with Phaseolus, the common bean plant, he encountered a number of complications. First, the hybrid plants were often sterile, making it difficult to collect enough F2 offspring. Second, some traits, for example, petal color, did not behave as discrete units of white or purple-red color. Instead, F2 progeny displayed a range of colors from purple-red to pale violet and white. Remarkably, rather than throwing up his hands in disgust, he admitted that his earlier thinking was too simple for this case and that there were going to be instances in which his simple rules of inheritance did not work. And then, he went one step further and proposed a possible reason for the complexity – that some traits may be governed by more than one discrete unit (or gene). And he was right! That is, multiple genes were independently segregating in the F2 bean progeny, creating a spectrum of colors, and not just the colors of the original parent plants. Mendel did what all good scientists do – he modified his thinking in the face of new information instead of rigidly adhering to his original idea. We will discuss traits encoded by multiple genes in the Frontiers section.
In addition to his choice of peas, Mendel was also lucky in another way. The seven traits he studied were all inherited independently of one another and thus he observed the 9:3:3:1 ratio in every combination of two traits he tested in F2 crosses. Today we know that this is because he had chosen traits encoded by genes that are either on separate chromosomes (or are sufficiently far apart on the same chromosome) that they behave independently in crosses. If he had chosen two traits that are "linked” – that is, close to one another on the same chromosome, they would have behaved as a single inheritance unit, and he would not have seen the independent assortment that was so important to his conclusions. For more information on gene linkage, see the Narrative on Plant Genetics by Ronald and the White Board Video on the Morgan and Sturtevant experiment.
Mendel presented his work publicly in Brno in 1865 and published it in the Brno Society for Natural History’s Proceedings in 1865. Unfortunately, Mendel’s publication received little attention and no one at the time understood its implications. It was not until 1900 – 16 years after Mendel’s death – that three European botanists – Hugo de Vries (1848–1935), Carl Correns (1864–1935), and Erich Tschermak (1871–1962) independently came upon Mendel’s paper in the course of conducting their own hybridization experiments and the import of Mendel’s work became evident.
While Mendel was completing his work, Darwin published his monumental treatise on evolution (Origin of the Species, 1859) but without the advantage of knowing about the laws of inheritance. Likewise, there is no compelling evidence that Mendel knew of Darwin’s thinking, at least at the time he performed his experiments. Can you imagine what a conversation between these two giants would have been like? Such a meeting would almost certainly have changed the history of biology! Scientists today appreciate the importance of publishing their work, not only to establish credit for their discoveries, but to allow others to rapidly build upon their findings, so we can all maximally learn more about how the natural world works.
It was only in 1910 that Thomas Hunt Morgan, working with the fruit fly Drosophila melanogaster at Columbia University, showed that genes, the word given for the unit of inheritance discovered by Mendel, reside on chromosomes that could be visualized in a microscope (see the Whiteboard Video on Morgan’s Discoveries). And it wasn’t until 1944 that three scientists working on bacteria at Rockefeller University, Oswald Avery, Colin Munro MacLeod, and Maclyn McCarty, demonstrated convincingly that genes are composed of deoxyribonucleic acid, DNA (see the White Board Video of the Avery, MacLeod, and McCarty Experiment). Finally, in 1953, James Watson and Francis Crick published their groundbreaking paper in which they proposed a double helical structure for DNA, which provided a clear answer to the important question of how a gene could be so faithfully replicated at each generation (see the Narrative on DNA Structure by Vale).
Part II: Knowledge Overview —
The Fundamentals of Inheritance
Mendel’s Laws
With his simple experiments, Mendel discovered the following general principles of inheritance:
- Simple traits are inherited from parents as single discrete units, which were later given the name “genes.”
- Each parent contributes to his or her offspring one copy of each gene in the production of gametes (pollen and ovules, in the case of plants; eggs or sperm in the case of animals). After fertilization, each offspring thus contains two copies of each gene.
- Genes provide the information for traits or phenotypes and can come in two varieties – dominant and recessive. When a dominant and recessive trait is inherited together, the phenotype associated with the dominant trait will prevail.
- Simple traits are inherited independently of one another (although as we will see below, this is not always true if the traits are encoded by genes that are near to one another (linked) on the same chromosome).
It took nearly 100 years until Mendel’s units of heredity were understood in terms of a real physical entity – the structure and sequence of deoxyribonucleic acid – DNA. See Video 2 for a review of the key terms in genetics and the organization of DNA in humans.
Genes and Mendel’s Laws
Genes In Brief
Today we know many additional properties of genes and their mode of inheritance. A gene is a linear segment of DNA encoding information that is copied by the cell into an RNA molecule. In most cases, the RNA (messenger RNA) is translated into a protein that plays a role in the cell (see the Central Dogma). In some cases, the RNA itself has a functional role. The information in a gene is embedded in the order of the four DNA subunits, called adenine (A), thymidine (T), cytosine (C), and guanine (G). The “code” is a triplet one so that each of 61 possible coding triplets of DNA (e.g., ACT, GCC) is read by the cellular machinery as one of 20 amino acids that make up proteins, and 3 triplets (TAG, TAA, TGA) are reserved as stop codons, a signal to the machinery that it has come to the end of the protein-coding information. The ways in which the proteins and RNAs carry out their activities are what give rise to the observable phenotypic traits of an organism.
Variants in genes (called alleles; Video 2) arise spontaneously due to errors in DNA replication or by exposure to environmental mutagens such as tar in tobacco or UV irradiation from the sun (see the Narrative on Mutations by Koshland). The diversity in the colors of pea petals or the shapes of pea seeds is the result of variations in DNA sequence of the genes that determine these particular traits.
Genes are organized into long circular (in the case of bacteria) or linear (in the case of all other organisms) molecules of DNA, with the genes arranged in a linear fashion along their length. Most organisms, such as humans, are diploid, meaning they contain two copies of each gene, one inherited from each parent. Humans, for example, have 23 pairs of chromosomes, which differ in size and the number of genes they contain. Single celled organisms like bacteria contain only one copy of each gene and are referred to as haploid.
During the formation of gametes, in a process referred to as meiosis, the chromosome pairs segregate away from each other so that each gamete contains only one copy of each, and thus are haploid. At fertilization, the chromosomes in the gametes combine to restore two copies of each in the embryo, which is now restored to a diploid state.
Some human traits are explained by “Mendelian Inheritance”
What is most remarkable about Mendel’s findings is their relevance to all organisms, including humans. Many human genes behave as single “Mendelian” traits, in which dominant and recessive alleles segregate in a ratio of 3:1. These genes can be studied in extended families, called pedigrees, that are akin to the crosses that Mendel performed in peas. At least 3550 single gene traits – referred to as Mendelian traits in honor of the founder – have been catalogued in a database called Mendelian Inheritance in Man (www.omim.org).
Examples of traits that display simple Mendelian inheritance include:
- Freckles
- Dimples
- Near sightedness
- Blood Rh factor (described as + or – in blood type)
- Cleft chin
- Lip size
A number of human diseases also display “Mendelian genetics.” These include:
- Cystic fibrosis
- Sickle cell anemia
- Hemophilia
- Phenylketonuria
- Huntington's disease
- Tay Sachs
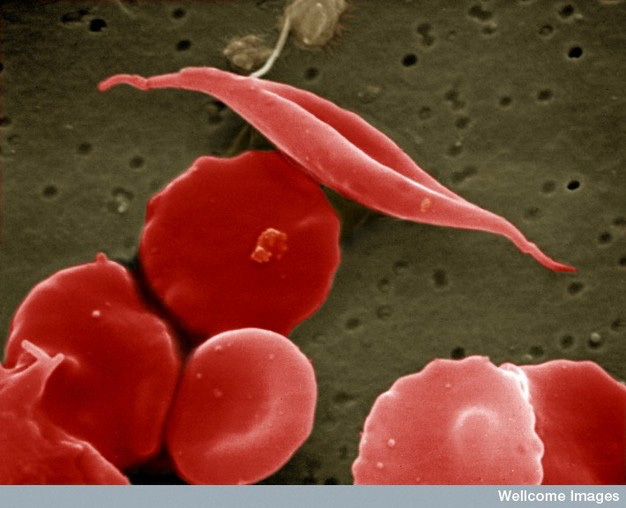
For example, the debilitating disease sickle cell anemia (SCA) is a relatively common genetic disease in the United States. The gene involved in SCA codes for hemoglobin, the protein in red blood cells that carries oxygen from the lungs to tissues throughout the body and returns carbon dioxide to the lungs. The mutation in sickle cell anemia is a miniscule change of a single DNA base in the hemoglobin gene from an adenine (A) to a thymine (T), which causes a change in one amino acid (from a glutamic acid to a valine; see the twenty types of amino acids) out of 146 amino acids in its polypeptide. Yet this one substitution is enough to alter the shape of the protein and its ability to bind oxygen. To make matters worse, the protein forms rigid aggregates that alter the overall shape of red blood cells, which is how the disease was named – some of the cells go from their normal spherical shape to looking like a crescent, or sickle (see Figure 7). Sickled cells do not navigate well through the small capillaries of the circulatory system and have a tendency to clump at their junctions, further inhibiting the delivery of oxygen to tissues.
The sickle cell trait behaves as a classical recessive Mendelian trait in families. As shown in the pedigree in Figure 8, an affected male in the second generation arises from a “cross” between parents who each carry one mutant (s) and one functional (S or wild type) copy of the hemoglobin gene (genotype – Ss). Statistically, they have a 1 in 4 chance of giving birth to a child who inherits the mutant version of the hemoglobin gene from both parents (ss). The parents are said to be carriers or heterozygous (one good copy and one mutant copy) for the sickle cell gene. The affected child is said to be homozygous (two mutant copies). If that affected individual grows up, marries a normal individual and has children, those children will become carriers of the disease (Ss).
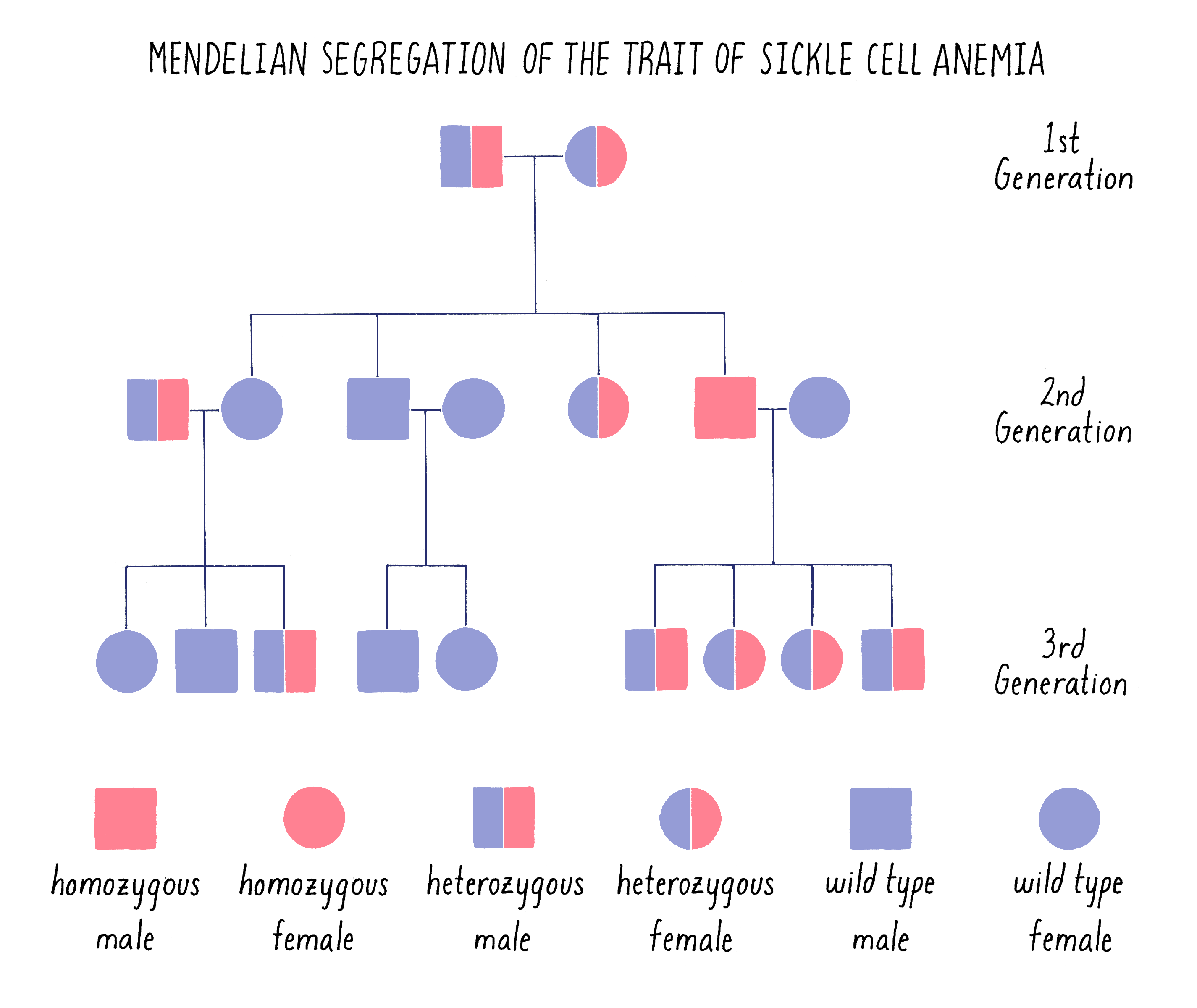
The pedigree in Figure 8 is drawn in the standard manner in which geneticists draw family trees, with males indicated by squares and females by circles. It should look very familiar to you after learning about Mendel and his peas.
Co-Dominant Traits
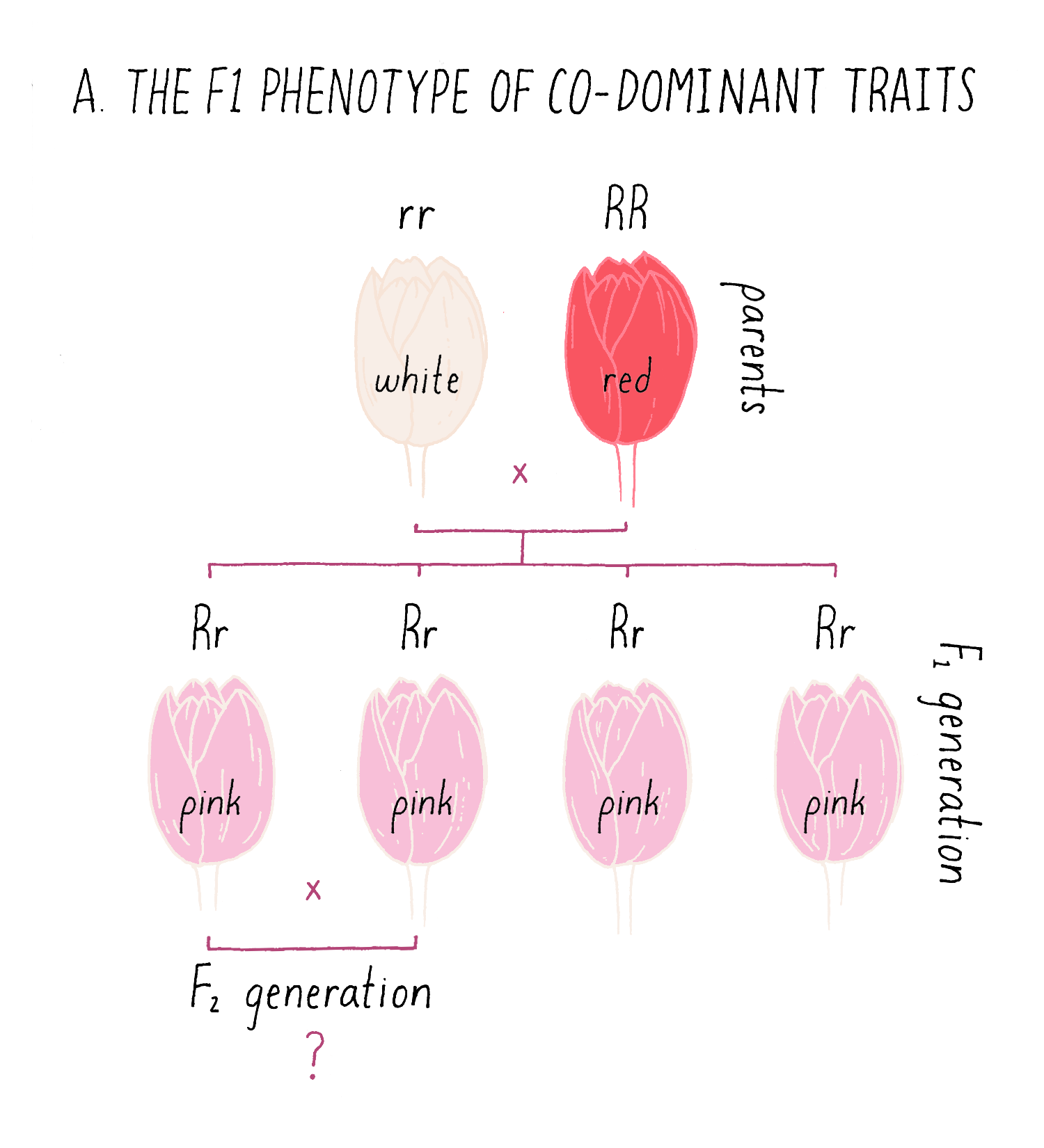
Although many traits observe the 3:1 inheritance rule that Mendel uncovered, some traits behave as co-dominant traits, in which the F1 phenotype is intermediate between the two parental types. In the example of a cross between white and red pigmented flowers in Figure 9A, the F1 plants all display a pink coloration, intermediate between the parental phenotypes.
Explorer’s Question: Can you predict the phenotypes of the progeny from of an intercross between two of the pink F1 plants (see the bottom of Figure 9A)?

Multi-Genetic Traits
Analogous to what Mendel discovered with the flowers of Phaseolus, most human traits are influenced by multiple genes. The genes responsible for these traits are much more difficult to study in families using pedigrees because they do not produce simple 3:1 inheritance patterns, but, rather, a spectrum of phenotypes like the flower pigmentation of Phaseolus.
Examples are multigenic traits include:
- Height
- Body shape
- Weight
- Intelligence
Examples of multigenic human diseases:
- Type II diabetes
- Coronary artery disease
- Alzheimer’s disease
- Schizophrenia
- Susceptibility to addiction
We will next explore how scientists are currently studying multigenic inheritance in the search for the underlying genes in the Frontiers section.
Part III: Frontiers —
Uncovering Genes Involved in Multi-Genetic Traits
The historic increase in lifespan in the United States has changed what people die from. One hundred years ago many people died of infectious diseases like smallpox, cholera, pneumonia, and tuberculosis. Many infectious diseases are to this day major challenges in developing countries. However, with the advent of better sanitation, vaccines, and antibiotics, the major causes of death in developed countries gradually switched to chronic illnesses such as coronary artery disease, type II diabetes, and cancer. In the future, it is predicted that dementias like Alzheimer’s will come to dominate the diseases of old age.
If we are going to tackle these maladies and increase the fraction of our normal lifespan when we can expect a high quality of life, we need to understand the underlying causes of these diseases. Why do Alzheimer’s and breast cancer run in some families? Knowing the genes that are involved may be the key to finding ways to counteract their deleterious effects.
The search for the genes that underlie the common multigenic diseases lies at the frontier of human genetics today (see Video 3: Whiteboard on Bioinformatics). The overall strategy is to search throughout the 3 billion DNA bases of the human genome and look for sequences that exist in common among those individuals with a certain trait, but differ from those who do not harbor that trait. Expressed in scientific terms, scientists are looking for identity by descent. For example, imagine a group of individuals who suffer from a very common disease like type II diabetes and a comparable group that does not. The modern geneticist is looking for statistical correlations between a particular DNA sequence in population A that is consistently different from that in population B. Given the size of the human genome, this task is like finding a needle in a haystack, or playing “where’s Waldo?”.
The sequencing of the human genome, completed in 2003, told us that humans are, on average 99.9% identical to one another in the order of their 3 billion bases of DNA. Yet the 0.1% difference translates into 3 million differences between any two individuals. The variations in single bases are called SNPs, for Single Nucleotide Polymorphisms, and they are scattered at random throughout the 23 pairs of chromosomes.
In some cases, the SNP might be meaningful, i.e., the change in sequence contributes to a phenotypic difference between individuals. An example is the SNP that changes an adenine (A) to a thymidine (T) in the hemoglobin gene and gives rise to sickle cell anemia (see the Knowledge Overview). However, since 98.5% of the human genome is noncoding (meaning that the sequence does not specify the sequence of a protein), the vast majority of SNPs are in noncoding regions. Even if the SNP is in a noncoding region, it could still affect the expression of a neighboring gene and thus change the levels or the timing of its protein product. In most cases, however, the SNP has no direct consequence for the individual. The change just arose by a spontaneous mutation (see the Narrative on by Mutations by Koshland) sometime in human history and was passed on from generation to generation. However, even if inconsequential, the SNPs are useful road signs, defining specific locations in the genome and its record of ancestry. By looking for particular SNPs, companies such as 23andMe and Ancestry.com can infer information about your ancestors (including whether some of them were likely to have been Neanderthals!). In the case of disease, the goal is to find one or more SNPs that correlate with the occurrence of a disease in a group of individuals. This kind of investigation is called a Genome-Wide Association Study (GWAS).
To illustrate how GWAS works, examine the frequencies of SNPs at two different positions in the genome in two populations – one population of individuals who have been diagnosed with diabetes and another population group free of diabetes. In the first example, SNP1, let’s say some individuals have an A at a specific location in the genome (GACTAT) as depicted by the purple man as opposed to a G and the same location (the green man; GGCTAT). The A SNP is found at a low frequency in both the diabetic group and the nondiabetic group (Figure 10A). Thus, there is no correlation between that SNP and the disease.
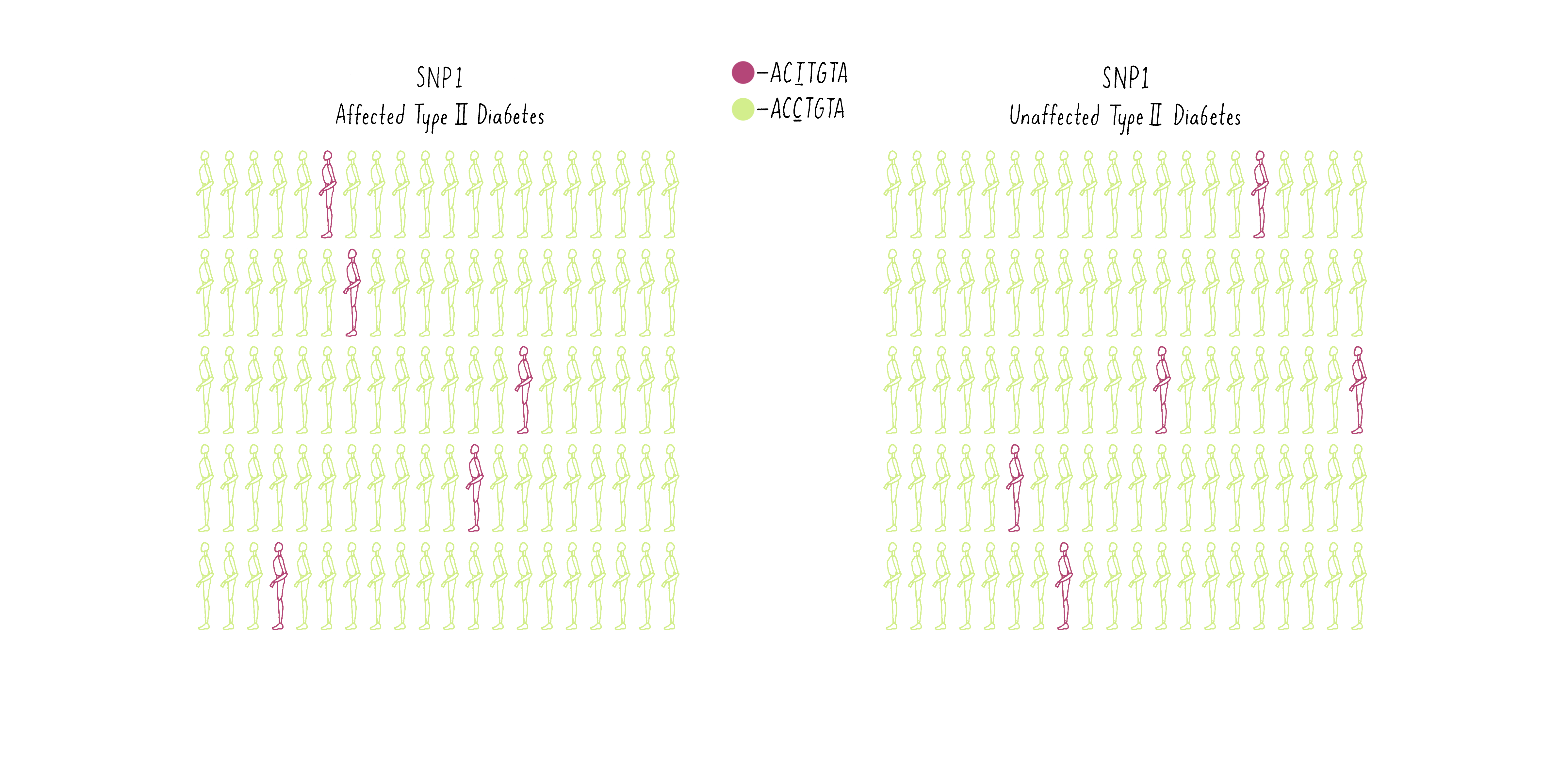
Now consider another SNP (SNP2 – let’s say a T at a different position in the genome (purple individuals; CCTGAG) as opposed to a C (green individuals; CCCGAG). The T is present at a higher frequency in the diabetic group than the population without diabetes (Figure 10B).
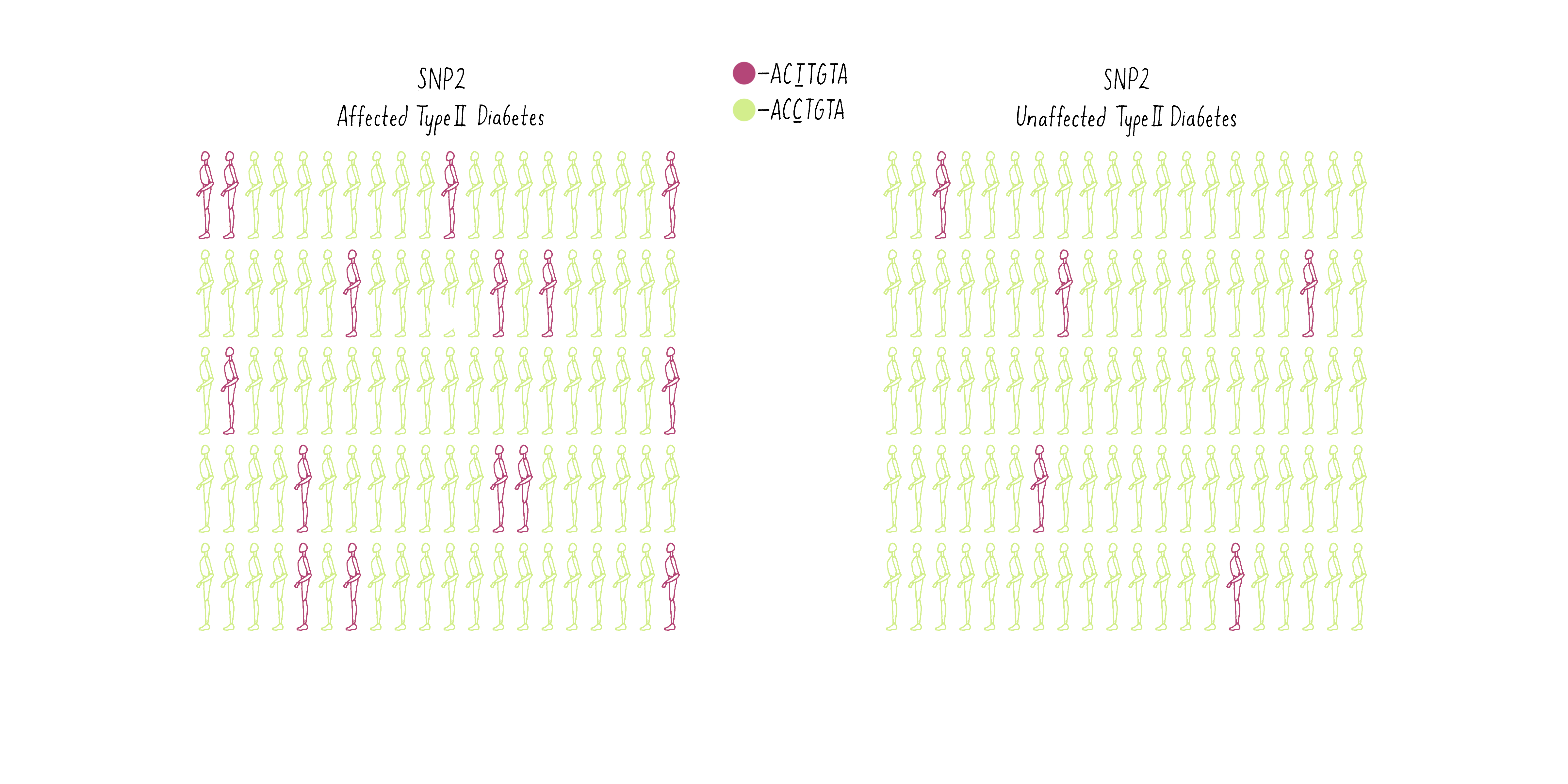
Of what value is the SNP to geneticists? Because the T allele of the SNP is more prevalent in the diabetic individuals, geneticists can conclude that a gene that contributes to the likelihood of becoming diabetic is most likely lurking close to the SNP. With this information, the gene search is no longer like “finding a needle in a haystack” but may come down to a few candidates, as we will discuss later.
What choices would a scientist make to set up such a GWAS? As with Mendel and his peas, choosing the right population to study is critical to the success of identifying traits influenced by multiple genes in humans. In 1996, an Icelandic scientist named Kari Stefansson founded a company called DeCODE Genetics to take advantage of the unique population in his island home.
Explorer’s Question: What makes the Icelandic population so ideal for GWAS?
- The island is home to a relatively homogeneous population of ~325,000, most of whom trace their heritage back to a small number of founders who settled the island around 875 AD.
- There has been very little immigration of new people to Iceland since then.
- Over the last 1140 years, there have been a number of natural catastrophes that reduced the population, creating “bottlenecks” in the population that further homogenized their genetic makeup.
- The Icelanders have kept very accurate genealogical records going back centuries.
- Iceland has a national health system that maintains comprehensive health records of all inhabitants.
- All of the above.
Answer: If you answered “All of the above,” you would be correct. With a relatively homogeneous population, the differences in DNA between those who exhibit a trait and those who do not are more likely to be related to the trait, rather than background “noise” due to random differences that have accumulated over millennia. Their extensive genealogical record keeping and accurate and comprehensive health records are enormously valuable to geneticists as they trace variant genes through a population and try to make correlations with disease.
Finally, studies of this kind require a cooperative group of volunteers who are willing to share their DNA and their health records with a private company. Not everyone would be so trusting, or willing to potentially risk their privacy. The company convinced the Icelandic people of the project by arguing that the high degree of relatedness of the population meant that their findings were likely to benefit not only the original DNA donors, but their relatives in the future.
In 2007, the scientists at DeCODE conducted a study of hair, eye, and skin pigmentation variation in a population of 2,986 Icelandic citizens. Choosing those traits was strategic, as there was prior evidence (including observations at family dining room tables) that those traits are influenced by multiple genes. They isolated DNA from each individual and determined the identity of the DNA SNPs (A, G, C, or T) at 317,511 positions in the genome that were known previously to be variable in human populations. They then conducted pairwise comparisons – blue eyes versus brown eyes, blond hair versus brown hair, freckles versus pale pigmentation, etc., looking for correlations between individual SNPs and the phenotypes of the individuals.
Even more than in Mendel’s experiments, the power of such studies to identify authentic correlations depends tremendously on the number of samples studied. The rarer the SNP or the rarer the trait, the more individuals that must be sampled. By conducting this study on a relatively homogeneous population like the Icelanders (fewer SNPs overall), the scientists had a much better chance that any differences they found would be significant and linked to the trait, and not due to random changes in DNA that accumulate over evolution.
This study identified 6 regions of the genome in which a SNP in a DNA base was correlated with specific eye, hair, or pigmentation phenotypes. For five of those regions, a specific gene was found near the SNPs that was a “smoking gun” candidate for playing a role in the particular trait. For example, one of the SNPs that was highly associated with light skin and sun sensitivity was located near the gene encoding the enzyme tyrosinase (TYR in Figure 11). This makes sense, since the mutations in tyrosinase are known to cause albinism (the lack of skin pigmentation) in humans. In another example, a SNP (T) that was associated with freckles was found near the MC1R gene. This gene encodes a protein receptor for melanocortin, a skin hormone.
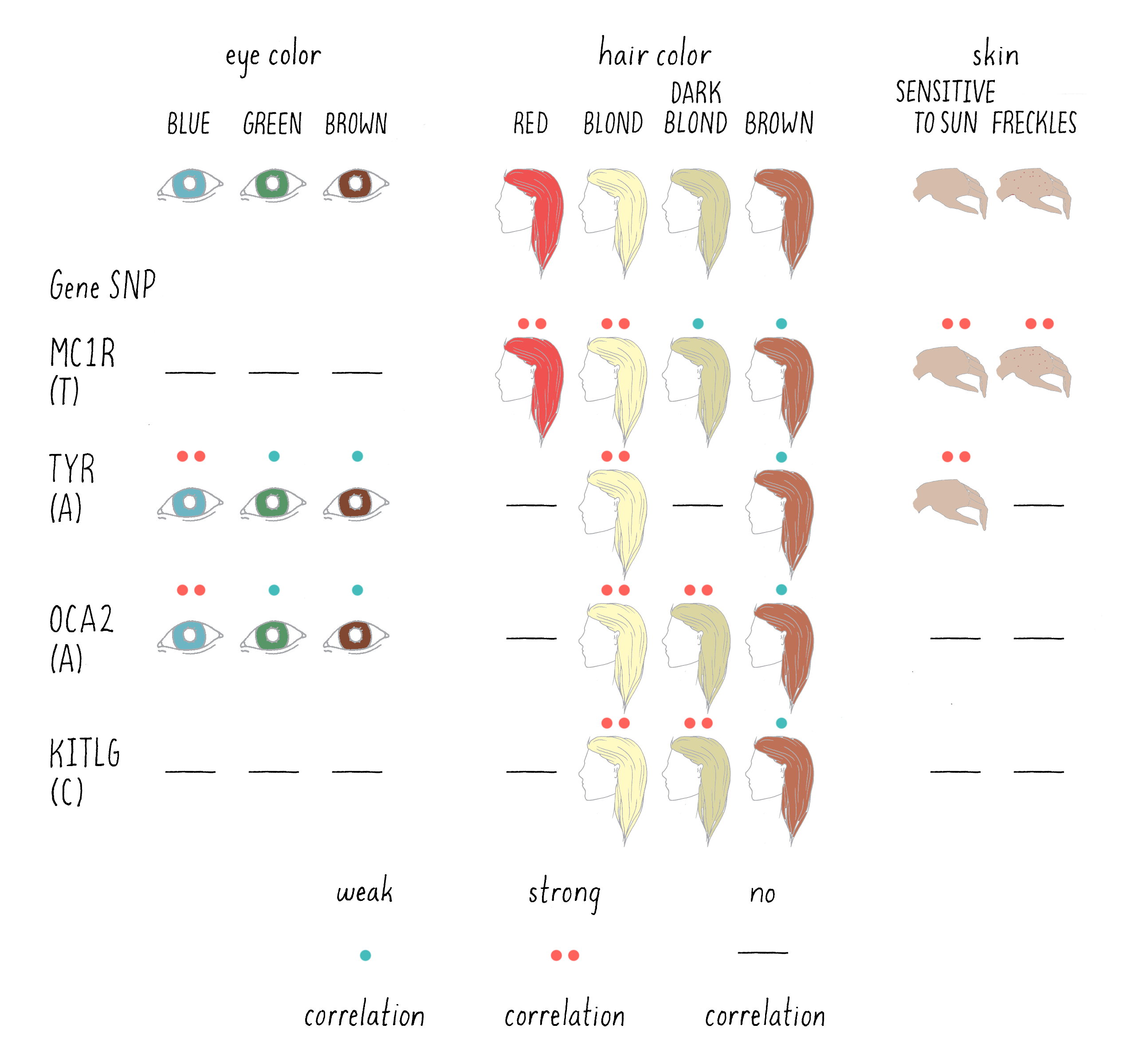
Of course, the correlations between SNP and trait were not perfect, even less so than Mendel’s ratios. For example, the SNP associated with MC1R was present in 13.5% of the total Icelandic sample population, but in 18.8% of those Icelandics with freckles. To persuade the scientists that the difference is real, and not the result of pure chance, statistical tests are needed. The statistics are expressed as probabilities (P) – defined as the likelihood that the association happened by chance. The larger the value of P, the greater the likelihood that the finding occurred by pure chance, with 1.0 representing complete certainty that the association is not significant. The smaller the number, the more confident one can be that the association is real and not due to chance.
A visual depiction of the DeCODE findings is presented in Figure 11 for the six genomic regions that showed a significant association with eye, skin, and hair pigmentation. A highly significant association is indicated by two asterisks (**) and a weak, but still a significant association is indicated by a single asterisk (*). A lack of a significant association is indicated by a dash (-).
For example, the trait of red hair was associated with the SNP near MC1R, with a probability of 4.2 × 10-95, an extraordinarily small number (meaning an extremely low probability that the SNP and phenotype were not connected). Thus, a scientist can conclude with great confidence that the MC1R gene is involved in determining red hair color. What do you associate with red hair? Freckles! And sure enough, the association with freckles and sensitivity to the sun are both associated with the same specific SNP near the MC1R gene(Figure 11). The A allele in the OCA2 gene, on the other hand, is more frequently found in those who are blond and have blue eyes. On the other hand, a SNP near to KITLG, a gene that encodes a hormone that binds to another type of cell surface receptor, had a P value of 0.84, meaning that it is very unlikely to affect red hair color.
Explorer’s Question:
- What traits are NOT associated with the MC1R gene?
- What trait is associated with a variant in only one gene?
- What traits are associated with the largest number of variants in genes?
- What traits are most significantly affected by the same gene?
- Eye color
- Red hair color
- Blond and brown hair
- Red hair, freckles, sensitivity to sun
One of the most powerful ways to assess the validity of a hypothesis is to test a prediction that it makes. In the case of genes and hair color, etc., one would predict that these correlations should be true of other populations, not just Iceland. The DeCODE scientists did just that by determining the SNP profile for the six genomic regions identified as important in the Icelandic sample in a completely unrelated set of 1,140 Dutch citizens. Using only the SNP information from the individuals, they predicted the color of eyes, hair, and skin of these individuals and then looked to see how well their predictions held up. To an amazing degree, the six SNPs were strong predictors of the Dutch phenotypes, as is evident from the similarity in the patterns of the two sets of bars for eye color in Figure 12. Of the 455 individuals who were predicted to have blue eyes, the vast majority of these individuals self-reported as being blue-eyed. Likewise, of the 210 individuals whom they predicted would be brown-eyed, most confirmed the prediction.
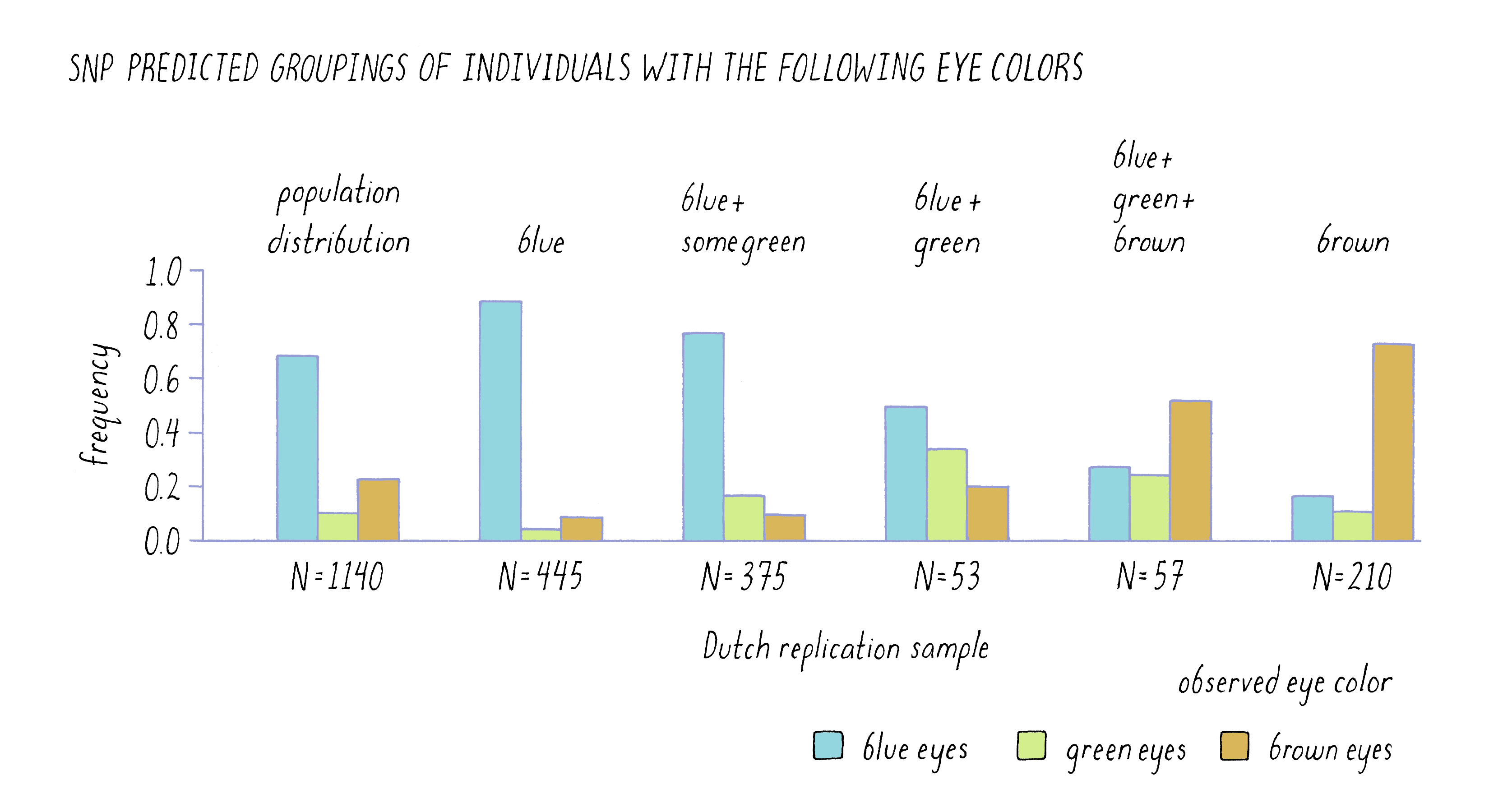
Their power to predict was not absolute, however. For example, they could not predict the eye color of 57 out of the 1,140 individuals (listed in the graph as blue, green, and brown which are all possible eye colors). When those individuals reported their eye color, they were a fairly even mixture of blue, brown, and green eyes. This result highlights the fact that although the scientists had identified the most important genes to affect eye color, there were likely to be other genes that had more modest effects individually but were acting collectively in those 57 individuals. Genes with modest effect or that work cooperatively in groups are very difficult to find in GWAS.
In summary, while there are many examples of simple Mendelian traits in humans, other traits and disease susceptibilities are influenced by the collective action of multiple genes. Those genes cannot be identified through classical Mendelian pedigree studies but can be identified by Genome-wide association studies (GWAS).
- GWAS exploit natural variation in the sequences of DNA in human populations – called single nucleotide polymorphisms (or SNPs) – as landmarks of specific regions in the genome.
- Large populations of affected and unaffected individuals are sampled for thousands of SNPs, and statistical correlations are sought between individual SNPs and phenotypes.
- The position of the SNP in the genome directs the scientists to the genes in the immediate locale that might be responsible for the phenotype.
Freckles are an interesting (and easier) proof-of-principle. But DeCODE and many other scientific groups around the world are on the hunt for the genes that underlie many diseases that show some evidence of a genetic component but do not display classical Mendelian inheritance. Many neurological and psychiatric diseases, such as schizophrenia, bipolar disease, and autism, fall into this category. Compounding the complexity of this genetic challenge, these diseases no doubt are influenced by experience and environmental factors. Such diseases and traits are known as multifactorial, meaning they have both genetic and environmental factors that influence their expression. However, if scientists can identify the collection of genes that increase the probability of developing these diseases, then perhaps they can understand the molecular defects that underlie these diseases. And with understanding comes hope – hope that new strategies (with drugs or even genetic engineering) might emerge to treat these diseases.
Closing Thoughts
Gregor Mendel’s experiments in his abbey’s garden are clearly among the most remarkable in the history of biology. Without any scientific colleagues with whom to share his results, Mendel must have been driven by his own curiosity. At a time when most naturalists were collecting specimens for museums, Mendel forged a new paradigm of experimentation and rigorous quantitative analysis. As I have highlighted in this Narrative, students (and senior scientists) today can learn a great deal about the scientific method by examining the choices that Mendel made a century and a half ago. Merely memorizing Mendel’s crosses and ratios is missing the beauty of his work. While Mendel’s findings were not appreciated in his lifetime, his work laid the foundation for a scientific revolution that continues to this day.
Every day scientists are using Mendel’s insights to explain natural phenomena in experimental organisms such as bacteria, fruit flies, and mice and to use that knowledge to better understand traits in ourselves. This is not easy! Most human traits are influenced by complex networks of many genes. To make matters more challenging for geneticists, the collective action of those genes is being modulated in each individual by environmental factors encountered both before and after birth. In other words, genes are not destiny; we are not hardwired by our genes, but are influenced by the complex interactions between many genes, and the environment in which we live.
Weight is an excellent example of a multifactorial trait that is affected by the complex interaction between genes and environment. There are rare mutations in humans that predispose individuals to obesity, but for the majority of the population, the risk of becoming overweight is determined by a large number of gene variants that interact with one another to affect metabolic rate, fat cell number, lipid biosynthesis, hunger, and satiety. Onto that genetic blueprint are imposed environmental factors, such as the nutritional status of the mother during pregnancy, the nature and quantity of food consumed after birth, and activity level. And it is not just the environment in the womb that can influence the outcome for fetuses. There is even recent evidence that the weight of a father can affect the likelihood of his offspring being overweight by nongenetic means, which is an example of the exploding field of epigenetics. Today, through GWAS, we know only a handful of the dozens if not hundreds of genes that affect weight, largely because most are likely to contribute only a very small amount to the outcome.
The major ongoing challenge for biomedical scientists today is to develop more precise and powerful ways to identify genes that influence specific human traits, to disentangle the contributions of genes and the environment to those traits, and to turn that knowledge into effective interventions that will ideally prevent and treat the multigenic diseases that are the major sources of both morbidity and mortality throughout the world. It is a grand challenge that Mendel would have relished. But today’s scientists are tackling these difficult problems in highly interactive, collaborative teams, rather than the isolation of an abbey. Through their efforts, we can expect great progress in the next twenty-five years in understanding the connections between genotype and phenotype in humans and many other species.
References and Resource
Guided Paper
Mendel G. 1866. Experiments in Plant Hybridization. Translated by Scott Abbott and Daniel J. Fairbanks. 2016. Genetics 204: 407–422. https://doi.org/10.1534/genetics.116.195198
This is a modern translation of the classic paper by Mendel. In addition to defining for the first time the concepts of dominant and recessive traits, Mendel’s research uncovered the laws of inheritance by which traits are passed from one generation to another.
DownloadReferences
- Sulem, P., Gudbjartsson, D.F., Stacey, S.N., et al. (2007). Genetic determinants of hair, eye, and skin pigmentation in Europeans. Nature Genetics 39: 1443–1452.
This paper by the DeCODE company using GWAS of 2986 Icelanders to identify genes involved in determining traits of hair, skin, and eye color. This study is featured in the Frontiers section.
Resource
- Punnett Square Calculator: http://scienceprimer.com/punnett-square-calculator
This webpage contains a two-factor Punnett square calculator as well as a description of what Punnett squares represent and how to solve the problems. In addition, there are extra sets of practice Punnett square problems and a video explaining Punnett squares.
- Central Dogma Video: https://www.dnalc.org/resources/3d/central-dogma.html
This short video describes the properties and relationships surrounding transcription and translation of proteins, often known as the “Central Dogma of Molecular Biology.”
- Sickle Cell Anemia Video: https://www.dnalc.org/resources/3d/17-sickle-cell.html
This short video provides an animation depicting the pathology of sickle cells anemia while describing the disease in detail.
- Mendelian Genetics, Probability, Pedigree and Chi-Square Statistics Activity: https://www.hhmi.org/biointeractive/mendelian-genetics-probability-pedigree-and-chi-square-statistics
This active learning activity requires students to work through a series of questions pertaining to the genetics of sickle cell anemia and its interesting connection to malaria. These questions will test students' comprehension of Mendelian genetics, probability, pedigree analysis, and chi-square statistics.
- Pedigrees and Inheritance of Lactose Intolerance: https://www.hhmi.org/biointeractive/pedigrees-and-inheritance-lactose-intolerance
This active learning activity walks students through some of the genetic changes associated with lactose tolerance/intolerance and how the trait is passed on in families.
- Dog Genomics (using GWAS) and Dogs as a Model Organism: https://www.hhmi.org/biointeractive/dog-genomics-and-dogs-model-organisms
This video features Dr. Elinor Karlsson explaining her GWAS in dogs and how she associates SNPs in genes with certain traits in dogs.
- Mapping Genes to Traits in Dogs Using SNPs: https://www.hhmi.org/biointeractive/mapping-genes-traits-dogs-using-snps
This activity shows students how to map specific SNPs that correlate to different traits in dogs.
Activity
Explore the beautiful shapes of pollen yourself with the Foldscope Activity on Pollen.